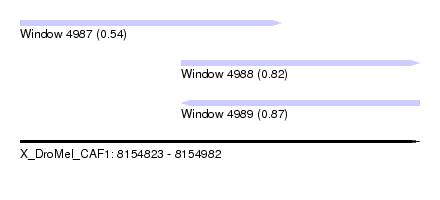
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,154,823 – 8,154,982 |
| Length | 159 |
| Max. P | 0.873725 |

| Location | 8,154,823 – 8,154,927 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -25.65 |
| Energy contribution | -25.74 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
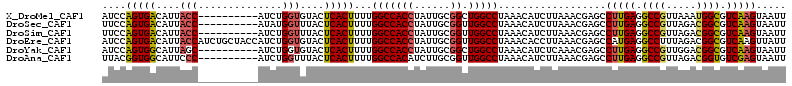
>X_DroMel_CAF1 8154823 104 + 22224390 AUCCAGUGACAUUACC----------AUCUGGUGUACUCACUUUUGGCCACCUAUUGCGGCUGGCCUAAACAUCUUAAACGAGCCUUGAGGCCGUUAAAUGGCGUCAAGUAAUU ....(((((...((((----------....))))...)))))...(((((((......)).)))))..................(((((.(((((...))))).)))))..... ( -32.40) >DroSec_CAF1 58057 104 + 1 UUCCAGUGACAUUACC----------AUAUGGUUUACUCACUUUUGGCCACCUAUUGCGGUUGGCCUAAACAUCUUAAACGAGCCUUGAGGCCGUUAGACGGCGUCAAGUAAUU ....(((((....(((----------....)))....)))))...(((((((......)).)))))..................(((((.((((.....)))).)))))..... ( -33.20) >DroSim_CAF1 39436 104 + 1 UUCCAGUGACAUUACC----------AUCUGGUUUACUCACUUUUGGCCACCUAUUGCGGUUGGCCUAAACAUCUUAAACGAGCCUUGAGGCCGUUAGACGGCGUCAAGUAAUU ....(((((....(((----------....)))....)))))...(((((((......)).)))))..................(((((.((((.....)))).)))))..... ( -33.20) >DroEre_CAF1 48819 114 + 1 AUCCAGUGACAUUACCAUCUGCUACCAUCUGGUGUACUCACUUUUGGCCACCUAUUGCGGUUGGCCUAAACACCUUAAACGAGCCAUGAGGCCUUUAGACGGCGUCAAGUUAUU ....((((((..........(((.......(((((..........(((((((......)).)))))...))))).......)))..(((.(((.......))).))).)))))) ( -30.26) >DroYak_CAF1 45884 104 + 1 AUCCAGUGGCAUUAGC----------AUCUGGUGUACUCACUUUUGGCCACCUAUUGCGGCUGGCCUAAACAUCUCAAACGAGCCUUGAGGCCGUUGGACGGCGUCAAGUAAUU ..((((..((....))----------..)))).............(((((((......)).))))).......(((....))).(((((.((((.....)))).)))))..... ( -33.30) >DroAna_CAF1 66862 104 + 1 UUACGGUGGCAUUCCC----------AUCUGGUUUACUCACUUUUGGCCACAUCUUGCGGUUGGCCUAAACAUCUUAAACGAGCCUUGAGGCCGUUAGACGGUGUCGAGUAAUU ....(((((.....))----------)))....((((((......(((((.(((....))))))))...(((((((.((((.(((....))))))).)).))))).)))))).. ( -33.40) >consensus AUCCAGUGACAUUACC__________AUCUGGUGUACUCACUUUUGGCCACCUAUUGCGGUUGGCCUAAACAUCUUAAACGAGCCUUGAGGCCGUUAGACGGCGUCAAGUAAUU ....(((((....(((..............)))....)))))...(((((((......)).)))))..................(((((.((((.....)))).)))))..... (-25.65 = -25.74 + 0.09)
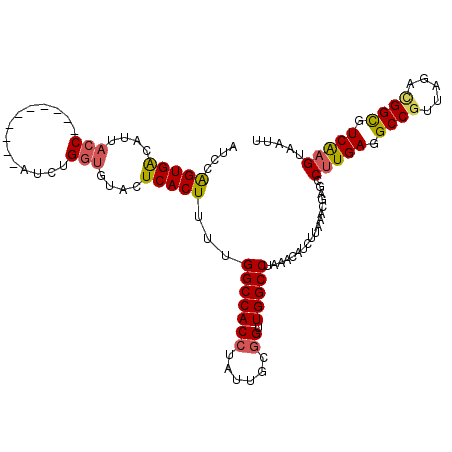
| Location | 8,154,887 – 8,154,982 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8154887 95 + 22224390 UUAAACGAGCCUUGAGGCCGUUAAAUGGCGUCAAGUAAUUUGGGCAAGCACAUGGCUGCUGGAUUCUGAAUUCCGAGUACACUCAUAAAAAGAUG .....((((.(((((.(((((...))))).)))))...))))((((.((.....))))))(((........)))(((....)))........... ( -24.90) >DroSec_CAF1 58121 94 + 1 UUAAACGAGCCUUGAGGCCGUUAGACGGCGUCAAGUAAUUUG-GCCAGCACAUGGCUGCUGGAUUCGGAAUUCCAAAUACACUCAUAAAAAGAUG ......(((.(((((.((((.....)))).)))))..(((((-(((((((......))))))..........))))))...)))........... ( -29.30) >DroSim_CAF1 39500 93 + 1 UUAAACGAGCCUUGAGGCCGUUAGACGGCGUCAAGUAAUUU--GCCAGCACAUGGCUGCUGGAUUCGGAAUUCCGAGUACACUCAUAAAAAGAUG ......(((.(((((.((((.....)))).)))))......--.((((((......))))))((((((....))))))...)))........... ( -33.50) >DroEre_CAF1 48893 94 + 1 UUAAACGAGCCAUGAGGCCUUUAGACGGCGUCAAGUUAUUUG-GACAGCACAUGGCUGCUGGAUUCCGAAUUCCGAGUACACUCAUAAAAAGAUG ............(((.(((.......))).))).(..(((((-(((((((......)))))...)))))))..)(((....)))........... ( -24.20) >DroYak_CAF1 45948 94 + 1 UCAAACGAGCCUUGAGGCCGUUGGACGGCGUCAAGUAAUUUG-GACAGCACAUGGCUGCUGGAUUCUGGAUUCCAACUGCACUUAUAAAAAAAUA ............((((((.((((((....(((.((.((((..-(.((((.....)))))..)))))).))))))))).)).)))).......... ( -26.70) >consensus UUAAACGAGCCUUGAGGCCGUUAGACGGCGUCAAGUAAUUUG_GCCAGCACAUGGCUGCUGGAUUCGGAAUUCCGAGUACACUCAUAAAAAGAUG ......(((.(((((.((((.....)))).)))))..........((((.....)))).((((........))))......)))........... (-21.42 = -21.50 + 0.08)


| Location | 8,154,887 – 8,154,982 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.62 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8154887 95 - 22224390 CAUCUUUUUAUGAGUGUACUCGGAAUUCAGAAUCCAGCAGCCAUGUGCUUGCCCAAAUUACUUGACGCCAUUUAACGGCCUCAAGGCUCGUUUAA .........((((((......(((........))).(((((.....)).)))........(((((.(((.......))).))))))))))).... ( -21.50) >DroSec_CAF1 58121 94 - 1 CAUCUUUUUAUGAGUGUAUUUGGAAUUCCGAAUCCAGCAGCCAUGUGCUGGC-CAAAUUACUUGACGCCGUCUAACGGCCUCAAGGCUCGUUUAA .........((((((..((((((....))))))((((((......)))))).-.......(((((.((((.....)))).))))))))))).... ( -31.40) >DroSim_CAF1 39500 93 - 1 CAUCUUUUUAUGAGUGUACUCGGAAUUCCGAAUCCAGCAGCCAUGUGCUGGC--AAAUUACUUGACGCCGUCUAACGGCCUCAAGGCUCGUUUAA .........((((((....((((....))))..((((((......)))))).--......(((((.((((.....)))).))))))))))).... ( -32.10) >DroEre_CAF1 48893 94 - 1 CAUCUUUUUAUGAGUGUACUCGGAAUUCGGAAUCCAGCAGCCAUGUGCUGUC-CAAAUAACUUGACGCCGUCUAAAGGCCUCAUGGCUCGUUUAA ...((((..(((.((((.(..(..(((.(((...(((((......)))))))-).)))..)..))))))))..))))(((....)))........ ( -21.80) >DroYak_CAF1 45948 94 - 1 UAUUUUUUUAUAAGUGCAGUUGGAAUCCAGAAUCCAGCAGCCAUGUGCUGUC-CAAAUUACUUGACGCCGUCCAACGGCCUCAAGGCUCGUUUGA ............((..((((((((........)))))).....))..))...-(((((..(((((.((((.....)))).)))))....))))). ( -26.60) >consensus CAUCUUUUUAUGAGUGUACUCGGAAUUCAGAAUCCAGCAGCCAUGUGCUGGC_CAAAUUACUUGACGCCGUCUAACGGCCUCAAGGCUCGUUUAA .........((((((..................((((((......)))))).........(((((.((((.....)))).))))))))))).... (-20.22 = -21.62 + 1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:45 2006