
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,133,879 – 8,134,020 |
| Length | 141 |
| Max. P | 0.999195 |

| Location | 8,133,879 – 8,133,980 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -15.69 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
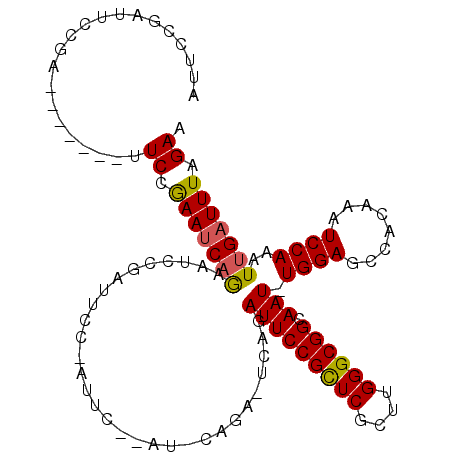
>X_DroMel_CAF1 8133879 101 + 22224390 AUGCCGAUUACCA-------AUCCGAAUCCGAUUUCGAUUCC------AAUUCAGAUUCCGAUUCCGUUCGCUUGGGCGGCAAUUUGGAGCCACAAAUCCAAAUUGAUUUAGAA ..((((....(((-------(..(((((..((((..((.((.------......)).)).))))..))))).)))).))))((((((((........))))))))......... ( -24.90) >DroEre_CAF1 24129 94 + 1 AUUGCGAUUCCGA-------UUCCAAAGCAGAAUGCGAUUCCAAUUC------------AGAUUCCGCUCGCUUGGGCGGCAAU-UGGAGCCACAAAUCCAAAUUGAUUUAGAA ....(((((..((-------((.....((.....))(..(((((((.------------.....((((((....)))))).)))-))))..)...))))..)))))........ ( -20.50) >DroYak_CAF1 24850 113 + 1 AUUCCGAUUCCGAUUCCCGAUUCCGAAUCAAAAUCCGAUUCCGAUUCCGAUCCAGAAUCGGAUUCCGCUCGCUUGGGCGGCAAU-UGGAGCCACAAAUCCAAAUUGAUUUAGAA ..((((((((.((((...((.((.(((((.......))))).)).)).))))..))))))))..((((((....))))))((((-((((........))).)))))........ ( -34.50) >consensus AUUCCGAUUCCGA_______UUCCGAAUCAGAAUCCGAUUCC_AUUC__AU_CAGA_UCAGAUUCCGCUCGCUUGGGCGGCAAU_UGGAGCCACAAAUCCAAAUUGAUUUAGAA .....................((.(((((((..............................(((((((((....)))))).))).((((........))))..))))))).)). (-15.69 = -15.70 + 0.01)

| Location | 8,133,912 – 8,134,020 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
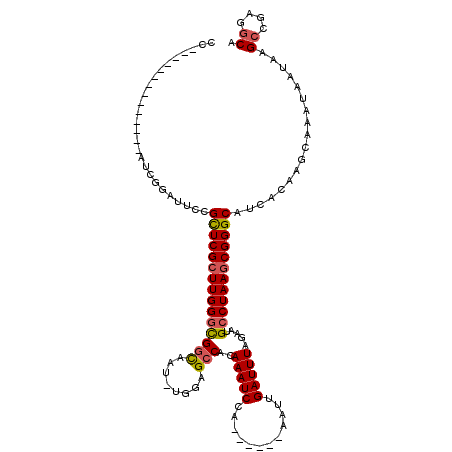
>X_DroMel_CAF1 8133912 108 + 22224390 CC------AAUUCAGAUUCCGAUUCCGUUCGCUUGGGCGGCAAUUUGGAGCCACAAAUCCA------AAUUGAUUUAGAAUGCCUAAGCGGGCAUCACAAGCAAAUAAUAAGCCGAGGCA ..------..................(((((((((((((.(((((((((........))))------)))))........)))))))))))))..................((....)). ( -34.40) >DroEre_CAF1 24162 101 + 1 CCAAUUC------------AGAUUCCGCUCGCUUGGGCGGCAAU-UGGAGCCACAAAUCCA------AAUUGAUUUAGAAUGCCUAAGCGGGCAUCACAAGCAAAUAAUAAGCCGGGGCA .......------------.(.(((((((((((((((((((...-....)))..(((((..------....))))).....)))))))))))).......((.........)).))))). ( -33.00) >DroYak_CAF1 24890 113 + 1 CCGAUUCCGAUCCAGAAUCGGAUUCCGCUCGCUUGGGCGGCAAU-UGGAGCCACAAAUCCA------AAUUGAUUUAGAAUGCCUAAGCGGGCAUCACAAGCAAAUAAUAAGCCGGGGCA (((((((.......))))))).....(((((((((((((((...-....)))..(((((..------....))))).....))))))))))))..................((....)). ( -41.50) >DroAna_CAF1 34523 103 + 1 ----------------AAAGCAUUCCGCUCGCUUGGCUGAUCAU-UCAAGCCACAAAUCCAAAGCAAAAUUGAUUUAAAAGGCCUAAACGGGCAUCACAACCAAAUAAUAAGCAAAGUCA ----------------..(((.....))).((((((((((....-)).)))).............................((((....))))................))))....... ( -17.40) >consensus CC______________AUCGGAUUCCGCUCGCUUGGGCGGCAAU_UGGAGCCACAAAUCCA______AAUUGAUUUAGAAUGCCUAAGCGGGCAUCACAAGCAAAUAAUAAGCCGAGGCA ..........................(((((((((((((((........)))..(((((............))))).....))))))))))))..................((....)). (-23.76 = -24.20 + 0.44)


| Location | 8,133,912 – 8,134,020 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.55 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
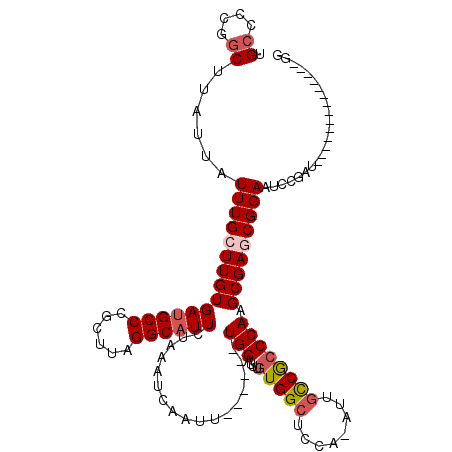
>X_DroMel_CAF1 8133912 108 - 22224390 UGCCUCGGCUUAUUAUUUGCUUGUGAUGCCCGCUUAGGCAUUCUAAAUCAAUU------UGGAUUUGUGGCUCCAAAUUGCCGCCCAAGCGAACGGAAUCGGAAUCUGAAUU------GG (((((.(((.((((((......))))))...))).)))))........(((((------..((((((((((........))))).....(((......))))))))..))))------). ( -33.20) >DroEre_CAF1 24162 101 - 1 UGCCCCGGCUUAUUAUUUGCUUGUGAUGCCCGCUUAGGCAUUCUAAAUCAAUU------UGGAUUUGUGGCUCCA-AUUGCCGCCCAAGCGAGCGGAAUCU------------GAAUUGG ....((((.(((...(((((((((((((((......))))))..........(------(((....(((((....-...))))))))))))))))))...)------------)).)))) ( -29.00) >DroYak_CAF1 24890 113 - 1 UGCCCCGGCUUAUUAUUUGCUUGUGAUGCCCGCUUAGGCAUUCUAAAUCAAUU------UGGAUUUGUGGCUCCA-AUUGCCGCCCAAGCGAGCGGAAUCCGAUUCUGGAUCGGAAUCGG .((....))......(((((((((((((((......))))))..........(------(((....(((((....-...))))))))))))))))))..(((((((((...))))))))) ( -39.20) >DroAna_CAF1 34523 103 - 1 UGACUUUGCUUAUUAUUUGGUUGUGAUGCCCGUUUAGGCCUUUUAAAUCAAUUUUGCUUUGGAUUUGUGGCUUGA-AUGAUCAGCCAAGCGAGCGGAAUGCUUU---------------- ....(((((((....((((((((.......((((((((((...((((((............)))))).)))))))-)))..)))))))).))))))).......---------------- ( -28.00) >consensus UGCCCCGGCUUAUUAUUUGCUUGUGAUGCCCGCUUAGGCAUUCUAAAUCAAUU______UGGAUUUGUGGCUCCA_AUUGCCGCCCAAGCGAGCGGAAUCCGAU______________GG .((....))......(((((((((((((((......)))))).................(((....(((((........)))))))).)))))))))....................... (-19.42 = -20.55 + 1.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:37 2006