
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,127,205 – 8,127,361 |
| Length | 156 |
| Max. P | 0.985448 |

| Location | 8,127,205 – 8,127,325 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
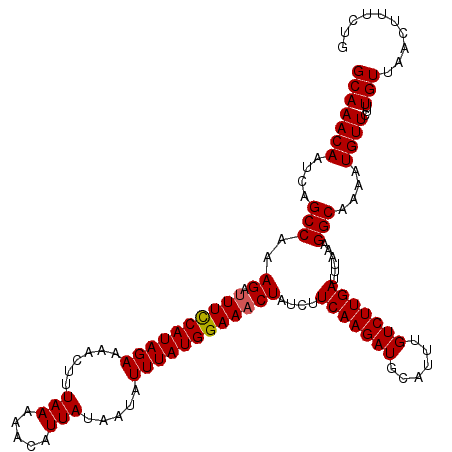
>X_DroMel_CAF1 8127205 120 + 22224390 GCAAACAAUCAGCCAAAGAUUUCCAUAGAAAACUUUAAAAACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUCUGUUAACUUUCUG (((((((....(((..((.((((((((((......(((.....))).....))))))))))))....(((((((......)))))))......)))....))))...))).......... ( -21.10) >DroSec_CAF1 30013 120 + 1 GCAAACAAUCAGCCAAAGAUUUCCAUAGAAAACUUUAAAAACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUUUGUUAACUUUCUG (((((.(((..(((..((.((((((((((......(((.....))).....))))))))))))....(((((((......)))))))......))).....))).))))).......... ( -21.10) >DroSim_CAF1 18715 120 + 1 GCAAACAAUCAGCCAAAGAUUUCCAUAGAAAACUUUAAAAACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUUUGUUAACUUUCUG (((((.(((..(((..((.((((((((((......(((.....))).....))))))))))))....(((((((......)))))))......))).....))).))))).......... ( -21.10) >DroEre_CAF1 17596 120 + 1 GCAAACAAUUAGCCAAAGACUUUCAUAGAAAACUUUAAAAACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGACUUAAAGGCCAAAUGUUCUUUGUUAACUUUCUG ........(((((.(((((.(((((((((......(((.....))).....)))))))))..............(((((((((((......)))).)))))))))))))))))....... ( -18.00) >DroYak_CAF1 17889 120 + 1 GCAAACAAUCAGCCAAAGAUUUUCAUAGAAAACUUUAAAAACAUUAUAAUAUUUAUGGAAACUAACUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCAUUGUUAACUUUCUG (((((((....(((..((.((((((((((......(((.....))).....))))))))))))....(((((((......)))))))......)))....)))...)))).......... ( -18.20) >consensus GCAAACAAUCAGCCAAAGAUUUCCAUAGAAAACUUUAAAAACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUUUGUUAACUUUCUG (((((((....(((..((.((((((((((......(((.....))).....))))))))))))....(((((((......)))))))......)))....))))...))).......... (-19.34 = -19.30 + -0.04)

| Location | 8,127,245 – 8,127,361 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8127245 116 + 22224390 ACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUCUGUUAACUUUCUGUGAGCUAUUGUUAUG----UAUGUAUGUAUGCAACUACAU ((((.((((((((((((((((.......(((((.(((((((((((......))))).))))))))).)).....)))))))))).)))))).)))----)(((((.((.....))))))) ( -24.80) >DroSec_CAF1 30053 112 + 1 ACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUUUGUUAACUUUCUGUGAGCUAUUGUUAU--------GUAUGUAUGCAACUACAU ((((.((((((((((((((((.......((((..(((((((((((......))))).))))))..)))).....)))))))))).)))))).))--------))(((((......))))) ( -25.60) >DroSim_CAF1 18755 120 + 1 ACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUUUGUUAACUUUCUGUGAGCUAUUGUUAUGUGUGUAUGUAUGUAUGCAACUACAU ((((.((((((((((((((((.......((((..(((((((((((......))))).))))))..)))).....)))))))))).)))))).))))(((((((((....))))..))))) ( -27.50) >DroEre_CAF1 17636 108 + 1 ACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGACUUAAAGGCCAAAUGUUCUUUGUUAACUUUCUGUGAGCUAUUGUUAUG--------U----AUGUAAGUACAU ((((.((((((((((((((((.......((((..(((((((((((......)))).)))))))..)))).....)))))))))).)))))).)))--------)----............ ( -24.30) >DroYak_CAF1 17929 108 + 1 ACAUUAUAAUAUUUAUGGAAACUAACUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCAUUGUUAACUUUCUGUGAGCUAUUGUUAUG--------U----ACGUAACUACGU ((((.((((((((((((((((.......(((...(((((((((((......))))).))))))...))).....)))))))))).)))))).)))--------)----((((....)))) ( -25.30) >consensus ACAUUAUAAUAUUUAUGGAAACUAUCUUCAAGAUGCAUUUGUCUUGAUUUAAAGGCAAAAUGUUCUUUGUUAACUUUCUGUGAGCUAUUGUUAUG_______GUAUGUAUGCAACUACAU .(((.((((((((((((((((.......((((..(((((((((((......))))).))))))..)))).....)))))))))).)))))).)))......................... (-20.56 = -21.16 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:35 2006