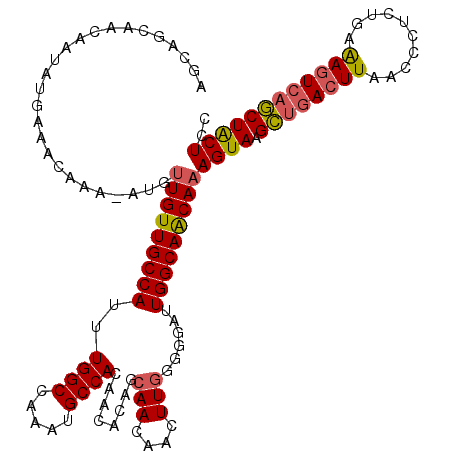
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,103,432 – 8,103,546 |
| Length | 114 |
| Max. P | 0.998149 |

| Location | 8,103,432 – 8,103,546 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8103432 114 + 22224390 -----CAACAAUAUGAAACAAA-AUGUUGUUGCCAUUUGGCCAAAUGCCACAGCAUAGCAACAACUUGGGGGAUUGGCAACAAAGUAAGCUGACUUAACUCUCUGAAAGUCAGCUACUCC -----.............(((.-.((((((((((....)))...((((....)))))))))))..)))((((.(((....)))....(((((((((..........))))))))).)))) ( -33.20) >DroSec_CAF1 8727 120 + 1 AGCAGCAACAAUAUGAAAAAAAAAAUUUGUUGCCAUUUGGCCAAAUGCCACAACACAGCAACAACUUGGGGGAUUGGCAACAAAGUAAGCUGACUUAACCCUCUGAAAGUCAGCUACUAC ..........................(((((((((..((((.....))))(..((.((......))))..)...)))))))))((((.((((((((..........)))))))))))).. ( -31.40) >DroSim_CAF1 14002 119 + 1 AGCAGCAACAAUAUGAAACAAA-AUGUUGUUGCCAUUUGGCCAAAUGCCACAACACAGCAACAACUUGGGGGAUUGGCAACAAAGUAAGCUGACUUAACCCUCUGAAAGUCAGCUACUAC ....((((((((((........-))))))))))......(((((.(.((.(((............))))).).))))).....((((.((((((((..........)))))))))))).. ( -35.50) >DroEre_CAF1 8257 117 + 1 GGCAGCAGCAAUAUGAAAUAAA-AUGCUGUUGCCAUUUGGCCAAAUGCCACAACACAGCAACAAGUUGG--GAUUGGCAACAAAGUAAGUUGACUUAACCCUCUGAGAGUCCACUGCUCU ((((((((((............-.)))))))))).....(((((...((.((((..........)))))--).))))).....((((.((.(((((..........))))).)))))).. ( -35.82) >DroYak_CAF1 11730 117 + 1 AGCAGCAACAAUAUGAAAUAAA-AUGUUGUUGCCAUUUGGCCAAAUGCCACAGCACAGCAACAACUUAG--GAUUGGCAGCAAAGUAAGUUUAGUUAACCCACUGAAAGUCAACUACUCC .((.((((((((((........-))))))))))....((((.....))))..))..............(--(((((((.....(((..(((.....)))..)))....)))))....))) ( -25.20) >consensus AGCAGCAACAAUAUGAAACAAA_AUGUUGUUGCCAUUUGGCCAAAUGCCACAACACAGCAACAACUUGGGGGAUUGGCAACAAAGUAAGCUGACUUAACCCUCUGAAAGUCAGCUACUCC ..........................(((((((((..((((.....))))........(((....)))......)))))))))((((.((((((((..........)))))))))))).. (-25.52 = -25.56 + 0.04)


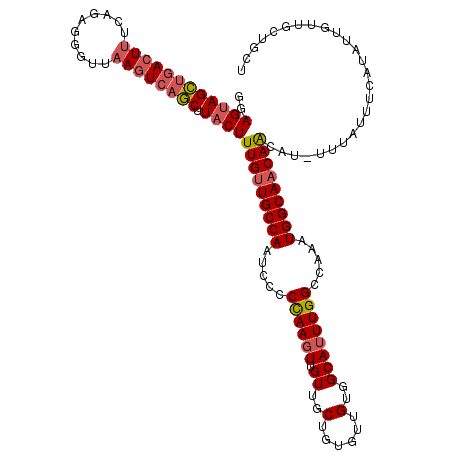
| Location | 8,103,432 – 8,103,546 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -30.48 |
| Energy contribution | -31.08 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8103432 114 - 22224390 GGAGUAGCUGACUUUCAGAGAGUUAAGUCAGCUUACUUUGUUGCCAAUCCCCCAAGUUGUUGCUAUGCUGUGGCAUUUGGCCAAAUGGCAACAACAU-UUUGUUUCAUAUUGUUG----- .(((((((((((((..........)))))))).)))))((((((((.....((((((.((..(......)..)))))))).....))))))))....-.................----- ( -37.70) >DroSec_CAF1 8727 120 - 1 GUAGUAGCUGACUUUCAGAGGGUUAAGUCAGCUUACUUUGUUGCCAAUCCCCCAAGUUGUUGCUGUGUUGUGGCAUUUGGCCAAAUGGCAACAAAUUUUUUUUUUCAUAUUGUUGCUGCU ((((((((((((((..........))))))))....((((((((((.....((((((.((..(......)..)))))))).....))))))))))..................)))))). ( -41.60) >DroSim_CAF1 14002 119 - 1 GUAGUAGCUGACUUUCAGAGGGUUAAGUCAGCUUACUUUGUUGCCAAUCCCCCAAGUUGUUGCUGUGUUGUGGCAUUUGGCCAAAUGGCAACAACAU-UUUGUUUCAUAUUGUUGCUGCU ((((((((((((((..........)))))))).....(((((((((.....((((((.((..(......)..)))))))).....)))))))))...-...............)))))). ( -40.70) >DroEre_CAF1 8257 117 - 1 AGAGCAGUGGACUCUCAGAGGGUUAAGUCAACUUACUUUGUUGCCAAUC--CCAACUUGUUGCUGUGUUGUGGCAUUUGGCCAAAUGGCAACAGCAU-UUUAUUUCAUAUUGCUGCUGCC ..(((((..(.........(((....(.((((.......)))))....)--))...((((((((((....((((.....)))).))))))))))...-...........)..)))))... ( -34.70) >DroYak_CAF1 11730 117 - 1 GGAGUAGUUGACUUUCAGUGGGUUAACUAAACUUACUUUGCUGCCAAUC--CUAAGUUGUUGCUGUGCUGUGGCAUUUGGCCAAAUGGCAACAACAU-UUUAUUUCAUAUUGUUGCUGCU ..((((((.(((....((((((((.....))))))))............--....(((((((((((....((((.....)))).)))))))))))..-.............))))))))) ( -32.90) >consensus GGAGUAGCUGACUUUCAGAGGGUUAAGUCAGCUUACUUUGUUGCCAAUCCCCCAAGUUGUUGCUGUGUUGUGGCAUUUGGCCAAAUGGCAACAACAU_UUUAUUUCAUAUUGUUGCUGCU ..((((((((((((..........)))))))).))))(((((((((.....((((((.((..(......)..)))))))).....))))))))).......................... (-30.48 = -31.08 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:23 2006