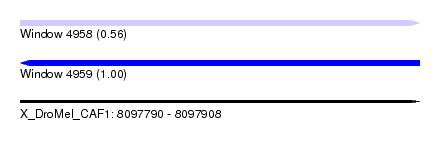
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,097,790 – 8,097,908 |
| Length | 118 |
| Max. P | 0.996917 |

| Location | 8,097,790 – 8,097,908 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -35.30 |
| Energy contribution | -36.54 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8097790 118 + 22224390 UGUUUCAUUUCUUCGGCGCGUGUAGAGUGGGCAAAUAUUAG--GCAUCAGAAGACUGAAAACUGGUUGCGACAACUUUCGUUGCGGCCGCCGAAGAAGUCGCCCCGUUUGGGCCAAGUUA ......(((((((((((...(((..((((......))))..--)))((((....)))).....(((((((((.......)))))))))))))))))))).((((.....))))....... ( -41.70) >DroSec_CAF1 3652 120 + 1 UGUUUCAUUUCUUCGGCGCGUGUAGAGUGGGCAAAUGUAAGGGGCCCCAGAAAACUGAAAACUGGUUGCGACAACUUUCGUUGCGGCCGCCGAAGAAGUCGCCCCGUUUGGGCCAAGUUA ......(((((((((((........(((((((...........))))(((....)))...)))(((((((((.......)))))))))))))))))))).((((.....))))....... ( -44.20) >DroSim_CAF1 8255 118 + 1 UGUUUCAUUUCUUCGGCGCGUGUAGAGUGGGCAAAUGUAAG--GCCCCAGAAGACUGAAAACUGGUUGCGACAACUUUCGUUGCGGCCGCCGAAGAAGUCGCCCCGUUUGGGCCAAGUUA ......(((((((((((........(((((((.........--))))(((....)))...)))(((((((((.......)))))))))))))))))))).((((.....))))....... ( -44.40) >DroEre_CAF1 3592 111 + 1 ---CUCACUUCAUUGGCGUGCA-AGAGUGGCAAA-UAUGAG--GC-CCAGAAGACUGAAAACUGGUUGCGACAACUUUCGUUGCGGCCGCCGAAGAAGUCGCC-CGUUUGGGCCAAGUUA ---...(((((.(((((.....-..((((((...-......--))-)(((....)))...)))(((((((((.......)))))))))))))).))))).(((-(....))))....... ( -37.50) >DroYak_CAF1 7055 117 + 1 UGUUUCAUUUCGCUGGCGUGUA-AGAGUGGGAAAUUAUGAG--GCCCCAGAAGACUGAAAACUGGUUGCGACAACUUUCGUUGCGGCCGCCGAAGAAGUCGCCCCGUUUGGGCCAAGUUA ....((((....((.((.....-...)).)).....))))(--((((.(((.((((......((((((((((.......)))))))))).......))))......))))))))...... ( -34.22) >consensus UGUUUCAUUUCUUCGGCGCGUGUAGAGUGGGCAAAUAUAAG__GCCCCAGAAGACUGAAAACUGGUUGCGACAACUUUCGUUGCGGCCGCCGAAGAAGUCGCCCCGUUUGGGCCAAGUUA ......(((((((((((........(((((((...........))))(((....)))...)))(((((((((.......)))))))))))))))))))).((((.....))))....... (-35.30 = -36.54 + 1.24)

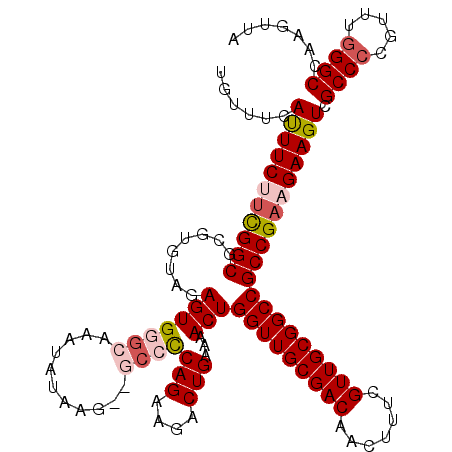

| Location | 8,097,790 – 8,097,908 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -34.14 |
| Energy contribution | -35.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8097790 118 - 22224390 UAACUUGGCCCAAACGGGGCGACUUCUUCGGCGGCCGCAACGAAAGUUGUCGCAACCAGUUUUCAGUCUUCUGAUGC--CUAAUAUUUGCCCACUCUACACGCGCCGAAGAAAUGAAACA .......((((.....))))...((((((((((((.(((((....))))).)).....((..((((....)))).))--.......................))))))))))........ ( -39.60) >DroSec_CAF1 3652 120 - 1 UAACUUGGCCCAAACGGGGCGACUUCUUCGGCGGCCGCAACGAAAGUUGUCGCAACCAGUUUUCAGUUUUCUGGGGCCCCUUACAUUUGCCCACUCUACACGCGCCGAAGAAAUGAAACA .......((((.....))))...((((((((((((.(((((....))))).))..........(((....)))((((...........))))..........))))))))))........ ( -43.30) >DroSim_CAF1 8255 118 - 1 UAACUUGGCCCAAACGGGGCGACUUCUUCGGCGGCCGCAACGAAAGUUGUCGCAACCAGUUUUCAGUCUUCUGGGGC--CUUACAUUUGCCCACUCUACACGCGCCGAAGAAAUGAAACA .......((((.....))))...((((((((((((.(((((....))))).))..........(((....)))((((--.........))))..........))))))))))........ ( -43.50) >DroEre_CAF1 3592 111 - 1 UAACUUGGCCCAAACG-GGCGACUUCUUCGGCGGCCGCAACGAAAGUUGUCGCAACCAGUUUUCAGUCUUCUGG-GC--CUCAUA-UUUGCCACUCU-UGCACGCCAAUGAAGUGAG--- .......((((....)-))).(((((...((((((.(((((....))))).))..((((...........))))-..--......-..(((......-.)))))))...)))))...--- ( -36.50) >DroYak_CAF1 7055 117 - 1 UAACUUGGCCCAAACGGGGCGACUUCUUCGGCGGCCGCAACGAAAGUUGUCGCAACCAGUUUUCAGUCUUCUGGGGC--CUCAUAAUUUCCCACUCU-UACACGCCAGCGAAAUGAAACA ......(((((...(((((.((((....(((..((.(((((....))))).))..)).).....)))))))))))))--).................-....((....)).......... ( -31.90) >consensus UAACUUGGCCCAAACGGGGCGACUUCUUCGGCGGCCGCAACGAAAGUUGUCGCAACCAGUUUUCAGUCUUCUGGGGC__CUCAUAUUUGCCCACUCUACACGCGCCGAAGAAAUGAAACA .......((((.....))))...((((((((((((.(((((....))))).))..((((...........))))............................))))))))))........ (-34.14 = -35.18 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:18 2006