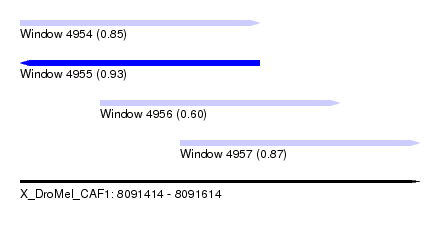
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,091,414 – 8,091,614 |
| Length | 200 |
| Max. P | 0.926607 |

| Location | 8,091,414 – 8,091,534 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -52.33 |
| Consensus MFE | -50.80 |
| Energy contribution | -52.30 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8091414 120 + 22224390 UGUGGCCUGUCUGGAUUUCGUUAAGCCCACGCCCACUCUGGGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCCCUUCU ...(((((((((((..........(((.(((((((((.(((...)))))).)))))).))).(((((..((....))..)).)))....)).))).((....))..))))))........ ( -51.80) >DroSim_CAF1 99 120 + 1 UGUGGCCUGUCUGGAUUACGUUAAGCCCACGCCCACUCUGGGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUUCU .(.(((((((((((..........(((.(((((((((.(((...)))))).)))))).))).(((((..((....))..)).)))....)).))).((....))..)))))).)...... ( -52.60) >DroYak_CAF1 119 120 + 1 UGUGGCCUGUCUGGAUUACGUUAAGCCCACGCCCACUCUGGGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUCCU .(.(((((((((((..........(((.(((((((((.(((...)))))).)))))).))).(((((..((....))..)).)))....)).))).((....))..)))))).)...... ( -52.60) >consensus UGUGGCCUGUCUGGAUUACGUUAAGCCCACGCCCACUCUGGGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUUCU .(.(((((((((((..........(((.(((((((((.(((...)))))).)))))).))).(((((..((....))..)).)))....)).))).((....))..)))))).)...... (-50.80 = -52.30 + 1.50)

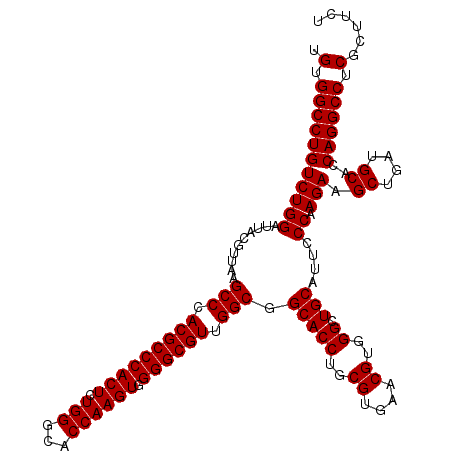

| Location | 8,091,414 – 8,091,534 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -52.03 |
| Consensus MFE | -51.30 |
| Energy contribution | -51.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8091414 120 - 22224390 AGAAGGGAGGCCUGGUGCAUCAGCUUCUUGGGAAUGCAGCCCACGUUCACGCAGGUGCCGCCAACGCCCCACUUGGUGCCCAGAGUGGGCGUGGGCUUAACGAAAUCCAGACAGGCCACA ........((((((.(((((...((.....)).)))))((((((((((((...((.((.(((((........)))))))))...))))))))))))...............))))))... ( -50.50) >DroSim_CAF1 99 120 - 1 AGAAGCGAGGCCUGGUGCAUCAGCUUCUUGGGAAUGCAGCCCACGUUCACGCAGGUGCCGCCAACGCCCCACUUGGUGCCCAGAGUGGGCGUGGGCUUAACGUAAUCCAGACAGGCCACA ........((((((..((....))......(((.((((((((((((((((...((.((.(((((........)))))))))...)))))))))))))....))).)))...))))))... ( -52.80) >DroYak_CAF1 119 120 - 1 AGGAGCGAGGCCUGGUGCAUCAGCUUCUUGGGAAUGCAGCCCACGUUCACGCAGGUGCCGCCAACGCCCCACUUGGUGCCCAGAGUGGGCGUGGGCUUAACGUAAUCCAGACAGGCCACA ........((((((..((....))......(((.((((((((((((((((...((.((.(((((........)))))))))...)))))))))))))....))).)))...))))))... ( -52.80) >consensus AGAAGCGAGGCCUGGUGCAUCAGCUUCUUGGGAAUGCAGCCCACGUUCACGCAGGUGCCGCCAACGCCCCACUUGGUGCCCAGAGUGGGCGUGGGCUUAACGUAAUCCAGACAGGCCACA ........((((((..((....))......(((.((((((((((((((((...((.((.(((((........)))))))))...)))))))))))))....))).)))...))))))... (-51.30 = -51.63 + 0.33)


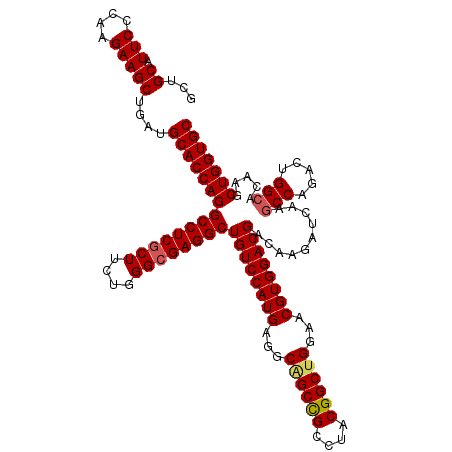
| Location | 8,091,454 – 8,091,574 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -57.83 |
| Consensus MFE | -53.53 |
| Energy contribution | -55.37 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8091454 120 + 22224390 GGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCCCUUCUGGGCGAGGCUGUCCAUGAGGCGGCCGCCUACGGCUGGAAC ....(((.(((((((((..((.(((((..((....))..)).))).(((....)))))..))))....((((..((((.((((((....)))))).)))).)))).)))))...)))... ( -55.80) >DroSim_CAF1 139 120 + 1 GGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUUCUGGGCGAGGCUGUCCAUGAGGCAGCUGCCUACGGCUGGAAC ....((..(((((((((..((.(((((..((....))..)).))).(((....)))))..))))....(((((((((....))))))))).)))))..))((((((....)))))).... ( -57.10) >DroYak_CAF1 159 120 + 1 GGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUCCUGGGCGAGGCUGUCCAUGAGGCUGCCGCCUACGGCUGGAAC ((((((..(((((((((..((.(((((..((....))..)).))).(((....)))))..))))....(((((((((....))))))))).)))))..)).))))(((...)))...... ( -60.60) >consensus GGCACCAAGUGGGGCGUUGGCGGCACCUGCGUGAACGUGGGCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUUCUGGGCGAGGCUGUCCAUGAGGCAGCCGCCUACGGCUGGAAC ....((..(((((((((..((.(((((..((....))..)).))).(((....)))))..))))....(((((((((....))))))))).)))))..))((((((....)))))).... (-53.53 = -55.37 + 1.84)



| Location | 8,091,494 – 8,091,614 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -50.50 |
| Consensus MFE | -48.36 |
| Energy contribution | -49.03 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8091494 120 + 22224390 GCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCCCUUCUGGGCGAGGCUGUCCAUGAGGCGGCCGCCUACGGCUGGAACGUGGACGAAAAGAUCAAGCCAGACUGGCACAAGCUGGUGC ...((.(((....)))))....((((((((((((((.....)).)))))((((((((...((((((....))))))...))))))))..........(((.....))).....))))))) ( -50.20) >DroSim_CAF1 179 120 + 1 GCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUUCUGGGCGAGGCUGUCCAUGAGGCAGCUGCCUACGGCUGGAACGUGGACGACAAGAUCAAGCCAGACUGGCACAAGCUGGUGC ...((.(((....)))))....(((((((((((((((....))))))))((((((((...((((((....))))))...))))))))..........(((.....))).....))))))) ( -52.60) >DroYak_CAF1 199 120 + 1 GCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUCCUGGGCGAGGCUGUCCAUGAGGCUGCCGCCUACGGCUGGAACGUGGACGACAAGAUCAAGCCAGACUGGAACAAACUGGUGC ...((.(((....)))))....(((((((((((((((....))))))))((((((..((((....))))..((((.((.(.((.....)).).)).))))....))).)))..))))))) ( -48.70) >consensus GCUGCAUUCCCAAGAAGCUGAUGCACCAGGCCUCGCUUCUGGGCGAGGCUGUCCAUGAGGCAGCCGCCUACGGCUGGAACGUGGACGACAAGAUCAAGCCAGACUGGCACAAGCUGGUGC ...((.(((....)))))....(((((((((((((((....))))))))((((((((...((((((....))))))...))))))))..........(((.....))).....))))))) (-48.36 = -49.03 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:16 2006