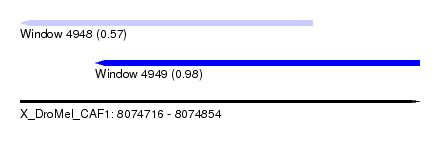
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,074,716 – 8,074,854 |
| Length | 138 |
| Max. P | 0.983053 |

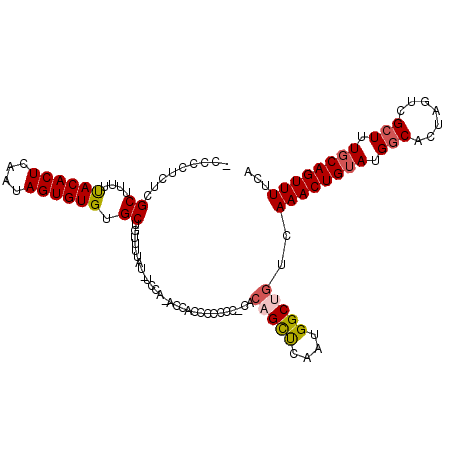
| Location | 8,074,716 – 8,074,817 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -18.46 |
| Consensus MFE | -12.34 |
| Energy contribution | -13.28 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8074716 101 - 22224390 UUUUUUUUUUCAUUUCCCCCCCGCACAGCUCAAUGGCUGUCAAACUGUAUGGCACUAGUCGCUUUGCAGUUUUCAAUUAAUUACAUUUAGCAAAUGUGUUU--- ......................(.((((((....)))))))((((((((.(((.......))).)))))))).........((((((.....))))))...--- ( -20.20) >DroSec_CAF1 56512 101 - 1 UUUUUCUUCACCUUCCCCCCACUCACAGCUCAAUGGCUGUCAAACUGUAUGGCACUAGUCGCUUUCCAGUUUUCAAUUAAUUACAUUUAGCAAAUGUGUUU--- ........................((((((....)))))).((((((...(((.......)))...)))))).........((((((.....))))))...--- ( -15.70) >DroEre_CAF1 43767 86 - 1 UUAAU--CCU-ACCA------------GCUCAAUGGCCGCCAAACUGUAUGACACCAGUCGCUUUGCAGUUUUCAAUUAAUUGCAUUUGGCACAUGUGUUU--- .....--...-.(((------------......)))..((((((......(((....)))....(((((((.......)))))))))))))..........--- ( -18.20) >DroYak_CAF1 51072 102 - 1 UUUACAGCCA-ACCACCCACCCACCCCGUCUAAUGGCUGUCAAACUGUAGGGCACUAGUCGCU-UGCAGUUUUCAAUUAAUUACAUUUAGCAAAUGUGUUUUUU ...(((((((-......................))))))).((((((((((.(.......)))-)))))))).........((((((.....))))))...... ( -19.75) >consensus UUUAU_UCCA_ACCACCCCCCC_CACAGCUCAAUGGCUGUCAAACUGUAUGGCACUAGUCGCUUUGCAGUUUUCAAUUAAUUACAUUUAGCAAAUGUGUUU___ .........................(((((....)))))..((((((((.(((.......))).)))))))).........((((((.....))))))...... (-12.34 = -13.28 + 0.94)

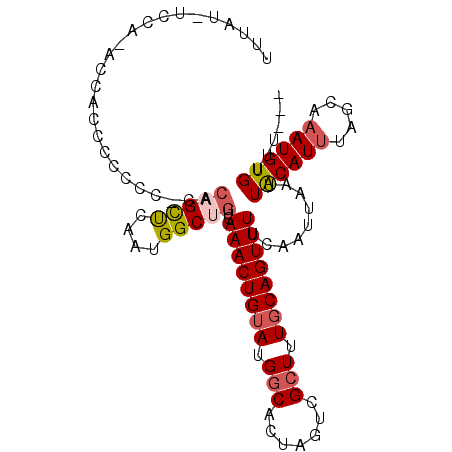
| Location | 8,074,742 – 8,074,854 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8074742 112 - 22224390 ACCCCACUCGCUUUUUACACUUAAUAGUGUGUGCUGUUUUUUUUUUUCAUUUCCCCCCCGCACAGCUCAAUGGCUGUCAAACUGUAUGGCACUAGUCGCUUUGCAGUUUUCA .........((....((((((....)))))).)).........................(.((((((....)))))))((((((((.(((.......))).))))))))... ( -25.00) >DroSec_CAF1 56538 112 - 1 ACCCCUCUCGCUUUUUACACUUAAUAGUGUGUGCUGUUUUUUCUUCACCUUCCCCCCACUCACAGCUCAAUGGCUGUCAAACUGUAUGGCACUAGUCGCUUUCCAGUUUUCA .........((....((((((....)))))).))...........................((((((....)))))).((((((...(((.......)))...))))))... ( -20.50) >DroEre_CAF1 43793 95 - 1 -AG-CUCUCGCUUUUUACACUCAAUAGUGUGUGCUGUUUAAU--CCU-ACCA------------GCUCAAUGGCCGCCAAACUGUAUGACACCAGUCGCUUUGCAGUUUUCA -.(-(....((....((((((....)))))).))........--...-.(((------------......)))..)).((((((((((((....))))...))))))))... ( -18.90) >DroYak_CAF1 51101 109 - 1 -CGCCUCUCGCUUUUCACACUCAACAGUGUGUGCAGUUUUACAGCCA-ACCACCCACCCACCCCGUCUAAUGGCUGUCAAACUGUAGGGCACUAGUCGCU-UGCAGUUUUCA -.((((.........((((((....))))))((((((((.(((((((-......................))))))).))))))))))))..........-........... ( -30.65) >consensus _CCCCUCUCGCUUUUUACACUCAAUAGUGUGUGCUGUUUUAU_UCCA_ACCACCCCCCC_CACAGCUCAAUGGCUGUCAAACUGUAUGGCACUAGUCGCUUUGCAGUUUUCA .........((....((((((....)))))).))............................(((((....)))))..((((((((.(((.......))).))))))))... (-18.14 = -18.83 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:09 2006