
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,064,468 – 8,064,603 |
| Length | 135 |
| Max. P | 0.986584 |

| Location | 8,064,468 – 8,064,583 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8064468 115 - 22224390 GGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUUAAAACACGGUGAUAAAAUAUCACUAAAUUGGUGCAUAUUCUUUA .....((((..((((.((((((......))).)))...))))..))))(((((.(((....((((((.((....))..(((((((...)))))))..)))))))))))))).... ( -28.80) >DroSec_CAF1 46422 99 - 1 GGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAAC---------------GGAGA-AAAAUAUCACUUAACUAGUGCAUUUUGUCUA ((((.((((..((((.((((((......))).)))...))))..))))......)))).....---------------(((.(-(((....((((.....))))..)))).))). ( -24.60) >DroSim_CAF1 22179 114 - 1 GGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUUAAAGCACGGUGA-AAAAUAUCACUUAACUAGUGCAUUUUGUCUA (((((((((.(((((...((((...((((((.(((.........))).).)))))))))...))))).))))..(((((((((-......)))))......))))...))))).. ( -30.90) >DroEre_CAF1 34242 110 - 1 GGCACUAGCAGUUGGAUCUGGUUUAAUUGCCCAGCAAACCAAAAUCUGGAGUAGUGCCAUAAUCGAUAGUUAAAGCACCGAUAAACAAUGCAUGUGUAUUA-----UUUAAGAUA (((((((((((((((((...))))))))))((((...........))))..)))))))...(((....((....)).....((((.(((((....))))).-----)))).))). ( -26.60) >DroYak_CAF1 40453 115 - 1 GGCGAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAGAGCUGUAGUAAUGCCAUAACCGAUAGUUAAAACACGCAGAAAAAAUGUCCCUAUACUAGCGCAUUUUAAAUA (((((((((..((((.((((((......))).)))...))))..))))).....))))...........((((((..(((((................)).)))..))))))... ( -24.59) >consensus GGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUUAAAACACGGAGA_AAAAUAUCACUAAACUAGUGCAUUUUAUAUA ((((.((((..((((.((((((......))).)))...))))..))))......))))......................................................... (-16.20 = -16.44 + 0.24)

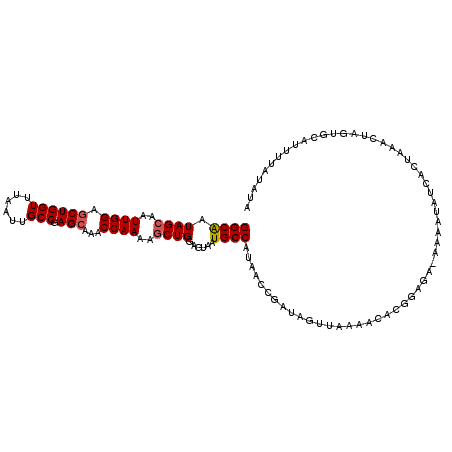
| Location | 8,064,508 – 8,064,603 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8064508 95 + 22224390 UUUUAACUAUCGGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCAUUUCCACUUUCGA ...........((...(((((.......(((..((((...(((.((........))))).))))..)))..)))))...)).............. ( -21.60) >DroSec_CAF1 46458 83 + 1 ------------GUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCAUUUCCCCUUUCAA ------------....(((((.......(((..((((...(((.((........))))).))))..)))..)))))................... ( -18.60) >DroSim_CAF1 22218 95 + 1 CUUUAACUAUCGGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCAUUUCCACUUUCAA ...........((...(((((.......(((..((((...(((.((........))))).))))..)))..)))))...)).............. ( -21.60) >DroEre_CAF1 34277 95 + 1 CUUUAACUAUCGAUUAUGGCACUACUCCAGAUUUUGGUUUGCUGGGCAAUUAAACCAGAUCCAACUGCUAGUGCCACUGCCAUUUCCACUUUCAA ................((((((((...(((...((((....((((.........))))..))))))).))))))))................... ( -22.50) >DroYak_CAF1 40493 95 + 1 UUUUAACUAUCGGUUAUGGCAUUACUACAGCUCUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUCGCCACUUCCAUUUCCACUUUCAA ...........((..((((.........(((..((((...(((.((........))))).))))..)))..........))))..))........ ( -19.31) >consensus CUUUAACUAUCGGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCAUUUCCACUUUCAA ...........((...(((((.......(((..((((...(((.((........))))).))))..)))..)))))...)).............. (-15.50 = -16.50 + 1.00)



| Location | 8,064,508 – 8,064,603 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8064508 95 - 22224390 UCGAAAGUGGAAAUGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUUAAAA .(....)......(((..((((((.((((..((((.((((((......))).)))...))))..))))......))))))..))).......... ( -23.90) >DroSec_CAF1 46458 83 - 1 UUGAAAGGGGAAAUGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAAC------------ ..................((((((.((((..((((.((((((......))).)))...))))..))))......))))))...------------ ( -20.90) >DroSim_CAF1 22218 95 - 1 UUGAAAGUGGAAAUGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUUAAAG ((((...(((..((((.....((..((((..((((.((((((......))).)))...))))..))))..))....))))..)))....)))).. ( -24.10) >DroEre_CAF1 34277 95 - 1 UUGAAAGUGGAAAUGGCAGUGGCACUAGCAGUUGGAUCUGGUUUAAUUGCCCAGCAAACCAAAAUCUGGAGUAGUGCCAUAAUCGAUAGUUAAAG .((...((....))..))(((((((((((((((((((...))))))))))((((...........))))..)))))))))............... ( -25.00) >DroYak_CAF1 40493 95 - 1 UUGAAAGUGGAAAUGGAAGUGGCGAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAGAGCUGUAGUAAUGCCAUAACCGAUAGUUAAAA ((((...(((..((((.....((.(((((..((((.((((((......))).)))...))))..))))).))....))))..)))....)))).. ( -26.40) >consensus UUGAAAGUGGAAAUGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUUAAAA .............(((..((((((.((((..((((.((((((......))).)))...))))..))))......))))))..))).......... (-19.36 = -19.84 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:04 2006