
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,966,183 – 7,966,330 |
| Length | 147 |
| Max. P | 0.968335 |

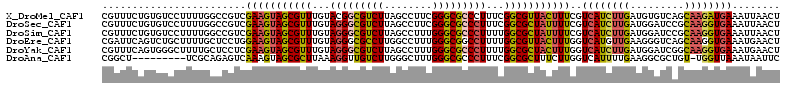
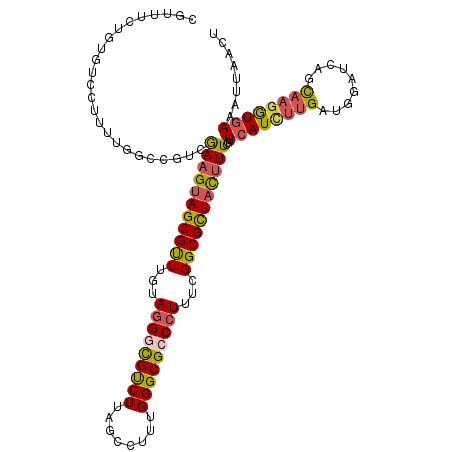
| Location | 7,966,183 – 7,966,296 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -32.24 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7966183 113 + 22224390 CGUUUCUGUGUCCUUUUGGCCGUCGAAGUAGCGUUUGUACGGCGUCUUAGCCUUCGGGCGCCCUUUCGGCGUUACUUUCGUCAUCUUGAUGUGUCAGCAAGAUGAAAUUAACU .........(.((....)).)(((((((..(((((((...(((......)))..)))))))..)))))))((((.(((((((...((((....))))...))))))).)))). ( -35.30) >DroSec_CAF1 3821 113 + 1 CGUUUCUGUGUCCUUUUGGCCGUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUCGGGCGCCCUUUCGGCGCUAUUUUCGUCAUCUUGAUGGAUCCGCAAGGUGAAAUUAACU .(((((.....((((.(((((((((((((((((((...(((((((((........)))))))))...))))))))).........)))))))..))).)))).)))))..... ( -41.40) >DroSim_CAF1 1809 113 + 1 CGUUUCUGUGUCCUUUUGGCCGUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUUGGGCGCCCUUUUGGCGCUAUUUUCGUCAUCUUGAUGGAUCCGCAAGGUGAAAUUAACU .(((((.....((((.(((((((((((((((((((...(((((((((........)))))))))...))))))))).........)))))))..))).)))).)))))..... ( -42.80) >DroEre_CAF1 1803 113 + 1 CGAUUCAGUCUGCUUUUGCUCCUGGAAGUAGCGUUUGUAGGGCGCCUUGGCCUUUGGGCGGCCUUUUGGCGUUACUUUGGUCAUGUUGAAGGGUCAGCAAGGUGAAAUGAACU .......(.((((((((......)))))))))(((..(....((((((((((....)))(((((((..((((.((....)).))))..)))))))..)))))))..)..))). ( -43.30) >DroYak_CAF1 1799 113 + 1 CGUUUCAGUGGGCUUUUGCUCCUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUUGGGCGCCCUUUUGGCGCUACUUUGGUCAUCUUGAUGGAUCGGCAAGGUGAAAUGAACU (((((((.(((((....))))..((((((((((((...(((((((((........)))))))))...))))))))))))(((((((....)))).)))..).))))))).... ( -50.40) >DroAna_CAF1 1847 103 + 1 CGGCU---------UCGCAGAGUCAAAGUAGCGCUUAAAGGUUGUCUUGGGCUUUGGGCGCCCUUUCGGCGCUUUCUUGGUCAUUUUGAAGGCGCUGU-UGGUUAAAUAAUUC .((((---------(....)))))......((((((((((.((.....)).))))))))))((...((((((((((...........)))))))))).-.))........... ( -35.70) >consensus CGUUUCUGUGUCCUUUUGGCCGUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUUGGGCGCCCUUUCGGCGCUACUUUCGUCAUCUUGAUGGAUCAGCAAGGUGAAAUUAACU ........................(((((((((((...(((((((((........)))))))))...)))))))))))..((((((((.........))))))))........ (-32.24 = -32.33 + 0.09)
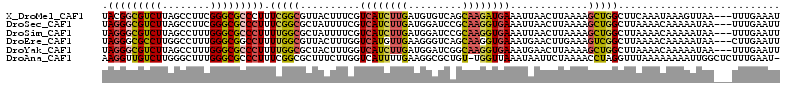
| Location | 7,966,220 – 7,966,330 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7966220 110 + 22224390 UACGGCGUCUUAGCCUUCGGGCGCCCUUUCGGCGUUACUUUCGUCAUCUUGAUGUGUCAGCAAGAUGAAAUUAACUUAAAAGCUGGCUUCAAAUAAAGUUAA---UUUGAAAU ..((((......(((....)))(((.....)))((((.(((((((...((((....))))...))))))).))))......))))..(((((((.......)---)))))).. ( -31.70) >DroSec_CAF1 3858 110 + 1 UAGGGCGUCUUAGCCUUCGGGCGCCCUUUCGGCGCUAUUUUCGUCAUCUUGAUGGAUCCGCAAGGUGAAAUUAACUUAAAAGCUGGCUUAAAACAAAAAUAA---UUUGAAUU .(((((((((........))))))))).(((((..........((((((((.((....)))))))))).............))))).......((((.....---)))).... ( -30.20) >DroSim_CAF1 1846 110 + 1 UAGGGCGUCUUAGCCUUUGGGCGCCCUUUUGGCGCUAUUUUCGUCAUCUUGAUGGAUCCGCAAGGUGAAAUUAACUUAAAAGCUGGCUUAAAACAAAAAUAA---UUUGAAUU .(((((((((........))))))))).(..((..........((((((((.((....)))))))))).............))..).......((((.....---)))).... ( -29.40) >DroEre_CAF1 1840 110 + 1 UAGGGCGCCUUGGCCUUUGGGCGGCCUUUUGGCGUUACUUUGGUCAUGUUGAAGGGUCAGCAAGGUGAAAUGAACUUGAAAGUCGGCUUAAAACAAAAAUAA---CUUGAAUU ((((.((((((((((....)))(((((((..((((.((....)).))))..)))))))..)))))))............(((....))).............---)))).... ( -34.20) >DroYak_CAF1 1836 110 + 1 UAGGGCGUCUUAGCCUUUGGGCGCCCUUUUGGCGCUACUUUGGUCAUCUUGAUGGAUCGGCAAGGUGAAAUGAACUUAAAAGCUGGCUUAAAACAAAAAUAA---UUUGAAUU .(((((((((........)))))))))(((((.((((((((..((((((((.........))))))))..........)))).))))))))).((((.....---)))).... ( -29.20) >DroAna_CAF1 1875 111 + 1 AAGGUUGUCUUGGGCUUUGGGCGCCCUUUCGGCGCUUUCUUGGUCAUUUUGAAGGCGCUGU-UGGUUAAAUAAUUCUAAAACCUAGGUUUAAAAAAAAUUGGCUCUUUGAAU- .(((((.....((((.......))))...((((((((((...........)))))))))).-.................)))))..(((((((............)))))))- ( -26.50) >consensus UAGGGCGUCUUAGCCUUUGGGCGCCCUUUCGGCGCUACUUUCGUCAUCUUGAUGGAUCAGCAAGGUGAAAUUAACUUAAAAGCUGGCUUAAAACAAAAAUAA___UUUGAAUU .(((((((((........))))))))).(((((..........((((((((.........)))))))).............)))))........................... (-19.84 = -20.23 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:33 2006