
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 974,990 – 975,086 |
| Length | 96 |
| Max. P | 0.796459 |

| Location | 974,990 – 975,086 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.09 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
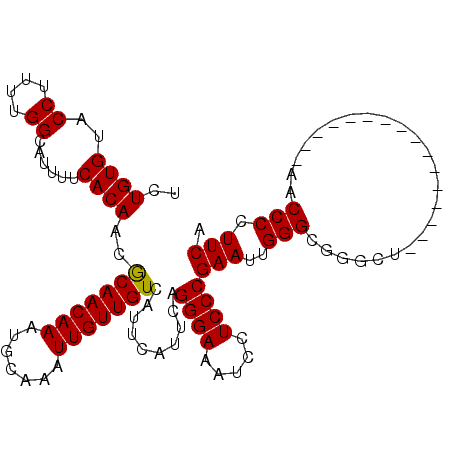
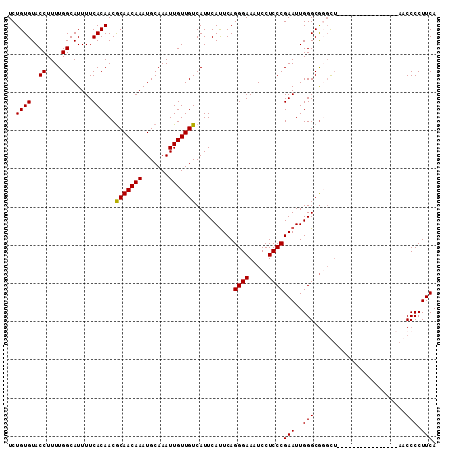
>X_DroMel_CAF1 974990 96 + 22224390 UCUGUGUACCUUUUGGCAUUUUCACAACGCAACAAAUGCAAAUUGUUGUCAUUCAUUCAGGGAAAUCCUCCCGAAUUGGGCGUGCU----------------ACCCCCUUCA .............((((((.((((....(((((((.......))))))).....((((.((((.....)))))))))))).)))))----------------)......... ( -20.00) >DroSec_CAF1 57947 96 + 1 UCUGUGUACCUUUUGGCAUUUUCACAACACAACAAAUGCAAAUUGUUGUCAUUCAUUCAGGGAAAUCCUCCCGAAUUGGGCAUGCU----------------AACCCCUUCA ............(((((((.((((....(((((((.......))))))).....((((.((((.....)))))))))))).)))))----------------))........ ( -20.30) >DroEre_CAF1 71240 102 + 1 UCUGUGUACCUUUUGGCAUUUUCACAACGCAACAAAUGCAAAUUGUUGUCAUUCACUCAGGGAAAUCCUCCCGAAUUGGGCGGGUUACGCC----------CAACCCCUUCA ..((((..((....))......))))..(((((((.......)))))))..........((((.....))))((((((((((.....))))----------)))....))). ( -26.60) >DroYak_CAF1 63330 112 + 1 UCUGUGUACCUUUUGGCAUUUUCACAACGCAACAAAUGCAAAUUGUUGUCAUUCAUUCAGGGAAAUCCUCCCGAAUUGGGCGGCUAUCCCCCUAACCCCUGCAACCCCUUCA ..((((..((....))......))))..(((((((.......)))))))..........((((.....))))(((..(((..((................))..))).))). ( -21.69) >consensus UCUGUGUACCUUUUGGCAUUUUCACAACGCAACAAAUGCAAAUUGUUGUCAUUCAUUCAGGGAAAUCCUCCCGAAUUGGGCGGGCU________________AACCCCUUCA ..((((..((....))......))))..(((((((.......)))))))..........((((.....))))(((..(((........................))).))). (-16.27 = -16.09 + -0.19)

| Location | 974,990 – 975,086 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -22.08 |
| Energy contribution | -21.71 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
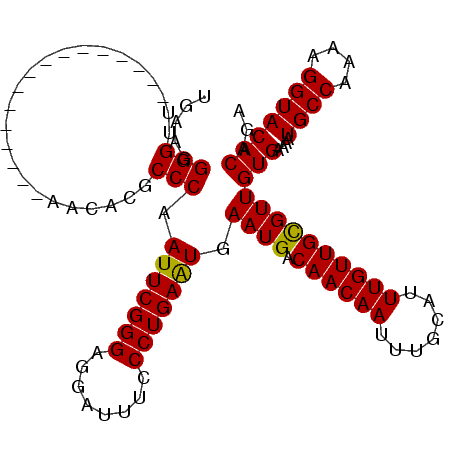
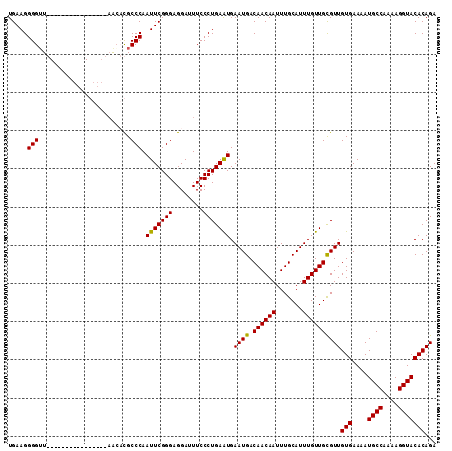
>X_DroMel_CAF1 974990 96 - 22224390 UGAAGGGGGU----------------AGCACGCCCAAUUCGGGAGGAUUUCCCUGAAUGAAUGACAACAAUUUGCAUUUGUUGCGUUGUGAAAAUGCCAAAAGGUACACAGA ....(((.(.----------------....).))).(((((((........))))))).((((.((((((.......))))))))))(((....((((....)))))))... ( -26.00) >DroSec_CAF1 57947 96 - 1 UGAAGGGGUU----------------AGCAUGCCCAAUUCGGGAGGAUUUCCCUGAAUGAAUGACAACAAUUUGCAUUUGUUGUGUUGUGAAAAUGCCAAAAGGUACACAGA .....((((.----------------.....)))).(((((((........))))))).....(((((((.......))))))).(((((....((((....))))))))). ( -26.10) >DroEre_CAF1 71240 102 - 1 UGAAGGGGUUG----------GGCGUAACCCGCCCAAUUCGGGAGGAUUUCCCUGAGUGAAUGACAACAAUUUGCAUUUGUUGCGUUGUGAAAAUGCCAAAAGGUACACAGA .....((((((----------((((.....))))))))))(((((...)))))...(((((((.((((((.......)))))))))).......((((....)))))))... ( -34.30) >DroYak_CAF1 63330 112 - 1 UGAAGGGGUUGCAGGGGUUAGGGGGAUAGCCGCCCAAUUCGGGAGGAUUUCCCUGAAUGAAUGACAACAAUUUGCAUUUGUUGCGUUGUGAAAAUGCCAAAAGGUACACAGA ..(((..((((((....((((((..((..((.(((.....))).))))..)))))).....)).))))..)))(((.....))).(((((....((((....))))))))). ( -34.10) >consensus UGAAGGGGUU________________AACACGCCCAAUUCGGGAGGAUUUCCCUGAAUGAAUGACAACAAUUUGCAUUUGUUGCGUUGUGAAAAUGCCAAAAGGUACACAGA .....(((........................))).(((((((........))))))).((((.((((((.......))))))))))(((....((((....)))))))... (-22.08 = -21.71 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:07 2006