
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,958,601 – 7,958,695 |
| Length | 94 |
| Max. P | 0.699954 |

| Location | 7,958,601 – 7,958,695 |
|---|---|
| Length | 94 |
| Sequences | 6 |
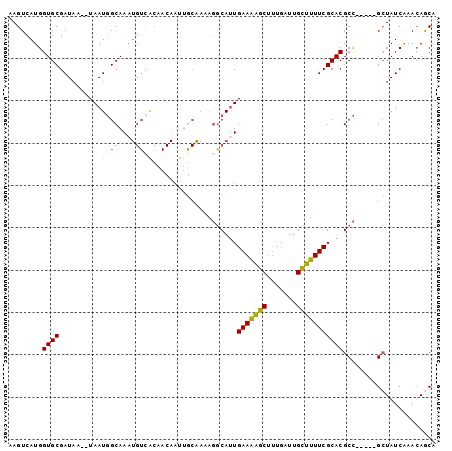
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7958601 94 + 22224390 AAGUCAUGGUGCGAU--UAUAAUGGCAAAUGUCACAACAAUUGCAAAAGGCAUUGAAAAACUUUGAUUGUUUUUCGCACGCC-----GCUAUCAAACAGCA .........((((((--(....((((....))))....)))))))...(((..((((((((.......))))))))...)))-----(((.......))). ( -22.50) >DroSec_CAF1 12346 96 + 1 AAGUCAUGGUGCGAUAAUAUAAUGGCAAAUGUCACAACAAUUGCAACGGGCAUUGAAAAGCUUUGAUUGCUUUUCGCACGCC-----GCUAUCGAACAGCA ........((.(((((........((((.(((....))).))))....(((..((((((((.......))))))))...)))-----..))))).)).... ( -24.60) >DroSim_CAF1 8622 96 + 1 AAGUCAUGGUGCGAUAAUAUAAUGGCAAAUGUCACAACAAUUGCAAAGGGCAUUGAAAAGCUUUGAUUGCUUUUCGCACGCC-----GCUAUCGAACAGCA ........((.(((((........((((.(((....))).))))....(((..((((((((.......))))))))...)))-----..))))).)).... ( -24.60) >DroEre_CAF1 9697 94 + 1 AAGUCAUGGUGCGAUAA--UAAUGGCAAAUGUCGCAACAACUGCGAAAGGCAUUGAAAAGCUUUGAUUGCUUUUCGCACGCC-----GCUAUCAAACAACA .......(((((((((.--..........))))))......((((((((((((..((....))..).))))))))))).)))-----.............. ( -25.80) >DroYak_CAF1 10021 76 + 1 AAGUCAUGGUGCGAUAA--UAAUGGCAA------------------AAGGCAUUGAAAGGCAUUGAUUGCUUUUCGCACGCC-----GCUAUCAAACAGCA ..(((((.((......)--).)))))..------------------..(((..(((((((((.....)))))))))...)))-----(((.......))). ( -21.60) >DroAna_CAF1 14691 98 + 1 AAGUCAUGGUGCGAUAA--UAAUUGCAAAUGUCACAACAACAACAACA-ACACUGAAGAGCUCUGAUUGCUUUUCGCACGCUGGCCUGAUAUCGAAAAACU ..((((..((((((((.--..........))))...............-.....(((((((.......)))))))))))..))))................ ( -19.10) >consensus AAGUCAUGGUGCGAUAA__UAAUGGCAAAUGUCACAACAAUUGCAAAAGGCAUUGAAAAGCUUUGAUUGCUUUUCGCACGCC_____GCUAUCAAACAGCA ........((((..........((((....))))....................(((((((.......)))))))))))...................... (-15.00 = -15.17 + 0.17)


| Location | 7,958,601 – 7,958,695 |
|---|---|
| Length | 94 |
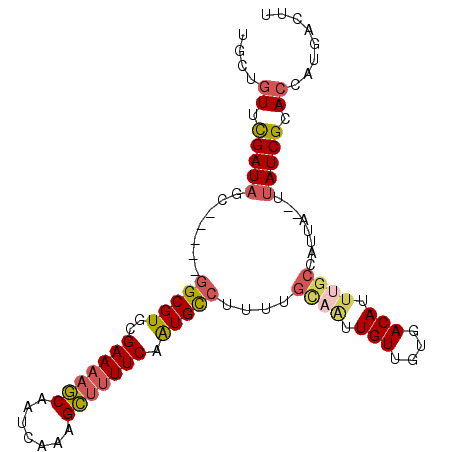
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7958601 94 - 22224390 UGCUGUUUGAUAGC-----GGCGUGCGAAAAACAAUCAAAGUUUUUCAAUGCCUUUUGCAAUUGUUGUGACAUUUGCCAUUAUA--AUCGCACCAUGACUU .((((((....)))-----)))(((((((((((.......)))))))..........((((.(((....))).)))).......--...))))........ ( -24.10) >DroSec_CAF1 12346 96 - 1 UGCUGUUCGAUAGC-----GGCGUGCGAAAAGCAAUCAAAGCUUUUCAAUGCCCGUUGCAAUUGUUGUGACAUUUGCCAUUAUAUUAUCGCACCAUGACUU ....((.((((((.-----(((((..(((((((.......))))))).)))))....((((.(((....))).)))).......)))))).))........ ( -26.00) >DroSim_CAF1 8622 96 - 1 UGCUGUUCGAUAGC-----GGCGUGCGAAAAGCAAUCAAAGCUUUUCAAUGCCCUUUGCAAUUGUUGUGACAUUUGCCAUUAUAUUAUCGCACCAUGACUU ....((.((((((.-----(((((..(((((((.......))))))).)))))....((((.(((....))).)))).......)))))).))........ ( -26.00) >DroEre_CAF1 9697 94 - 1 UGUUGUUUGAUAGC-----GGCGUGCGAAAAGCAAUCAAAGCUUUUCAAUGCCUUUCGCAGUUGUUGCGACAUUUGCCAUUA--UUAUCGCACCAUGACUU (((.((.((((.((-----(((((..(((((((.......))))))).)))))..((((((...)))))).....)).))))--..)).)))......... ( -25.10) >DroYak_CAF1 10021 76 - 1 UGCUGUUUGAUAGC-----GGCGUGCGAAAAGCAAUCAAUGCCUUUCAAUGCCUU------------------UUGCCAUUA--UUAUCGCACCAUGACUU .((((((....)))-----)))((((((((.(((.....))).))))((((.(..------------------..).)))).--.....))))........ ( -19.00) >DroAna_CAF1 14691 98 - 1 AGUUUUUCGAUAUCAGGCCAGCGUGCGAAAAGCAAUCAGAGCUCUUCAGUGU-UGUUGUUGUUGUUGUGACAUUUGCAAUUA--UUAUCGCACCAUGACUU ..............(((.((..((((((..((((((.(.(((........))-).).))))))((((..(...)..))))..--...))))))..)).))) ( -18.80) >consensus UGCUGUUCGAUAGC_____GGCGUGCGAAAAGCAAUCAAAGCUUUUCAAUGCCUUUUGCAAUUGUUGUGACAUUUGCCAUUA__UUAUCGCACCAUGACUU ....((.(((((.......(((((..(((((((.......))))))).)))))....((((.(((....))).))))........))))).))........ (-17.64 = -18.35 + 0.71)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:27 2006