
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,927,809 – 7,927,940 |
| Length | 131 |
| Max. P | 0.945111 |

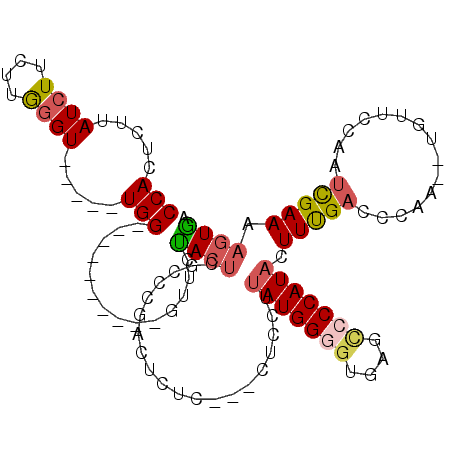
| Location | 7,927,809 – 7,927,902 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.62 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -9.04 |
| Energy contribution | -9.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7927809 93 - 22224390 CCAUUCUUAUCUUCAUGGGU-----UGG----------GCUCUACUCGCCCGACUCUC------UAUAUGGUGAGAGUCCCAUACUUUGACCCAA--UGUUCCAAUCGAAAAAUAA ...............(((((-----(((----------((.......))))(((((((------........))))))).........)))))).--................... ( -26.90) >DroSec_CAF1 3913 97 - 1 CCACUCUUAUCUUCUUGGGU-----UGG----------GUUCCACUCGCCCGACUCUC--GCUCCUUAUGGGGUGAGCCCCAUACUUUGACCCAA--UGUUCCAAUCGAAAAGUGA .((((.........((((((-----(((----------((.......))))....(((--(((((....))))))))...........)))))))--..(((.....))).)))). ( -28.50) >DroSim_CAF1 3063 99 - 1 CCACUCUUAUCUUCUUGGGU-----UGG----------GUUCCACUCACCCGACUCUCUCUCUCCUUAUGGGGUGAGCCCCAUACUUUGACCCAA--UGUUCCAAUCGAAAAGUGA .((((.........((((((-----(((----------((.......))))...............(((((((....)))))))....)))))))--..(((.....))).)))). ( -27.20) >DroEre_CAF1 4050 92 - 1 CCACUCCUAUCUUCUUAAGUU----UGG----------GUUCUACUGGGACGA--------UAUGGUAUGGGUUCAGCGCCAUACUUCGACCCAA--UCCUCCAACUGAAAAGUGA .((((((((.((.....))..----)))----------)......((((.(((--------...((((((((.....).)))))))))).)))).--..............)))). ( -22.10) >DroYak_CAF1 4225 108 - 1 CCACUCUUAUCUCCUUGGGUUGGGUUGGGUUCUACUUGGUUCUACUGCGUCCA--------CUCCUCAUGGGGUGAGCCCCAUACUUUGACCUCAUAUAUUCCAAUCGAAAUGUGA .................(((..(((((((.........((......))(..((--------((((....))))))..))))).)).)..)))((((((.(((.....))))))))) ( -25.80) >consensus CCACUCUUAUCUUCUUGGGU_____UGG__________GUUCUACUCGCCCGACUCUC___CUCCUUAUGGGGUGAGCCCCAUACUUUGACCCAA__UGUUCCAAUCGAAAAGUGA (((.....((((....)))).....)))..............((((....................(((((((....))))))).(((((...............))))).)))). ( -9.04 = -9.64 + 0.60)


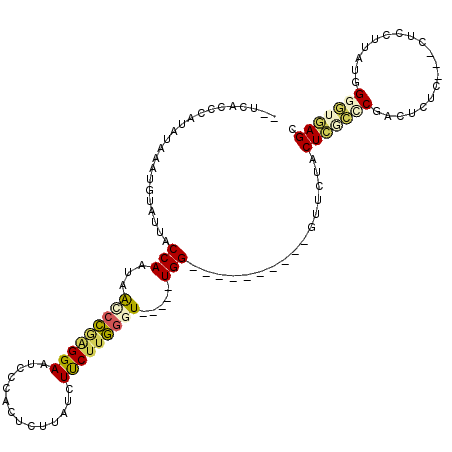
| Location | 7,927,845 – 7,927,940 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.51 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -11.49 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7927845 95 - 22224390 --UCACUCAUAUAAAUGUAUUACCAAUAACCCAAGGAAUCCCAUUCUUAUCUUCAUGGGU-----UGG----------GCUCUACUCGCCCGACUCUC------UAUAUGGUGAGAGU --((((.((((((....................((((((...))))))........((((-----(((----------((.......)))))))))..------)))))))))).... ( -25.80) >DroSec_CAF1 3949 99 - 1 --UCACCCAUAUGGAUGUAUUACCAAUGAGCCGAGGAAACCCACUCUUAUCUUCUUGGGU-----UGG----------GUUCCACUCGCCCGACUCUC--GCUCCUUAUGGGGUGAGC --((((((...(((........)))..((((.(((........)))..........((((-----(((----------((.......)))))))))..--))))......)))))).. ( -33.20) >DroSim_CAF1 3099 101 - 1 --UCACCCAUAUACAUGUAUUACCAAUGAGCCGAGGAAACCCACUCUUAUCUUCUUGGGU-----UGG----------GUUCCACUCACCCGACUCUCUCUCUCCUUAUGGGGUGAGC --((((((.((((.........((((.(((..(((........)))....))).))))((-----(((----------((.......)))))))............)))))))))).. ( -28.10) >DroEre_CAF1 4086 96 - 1 UGGCACCUUUAUACAUGUAUUACCAACAGCCUUGGGAAUCCCACUCCUAUCUUCUUAAGUU----UGG----------GUUCUACUGGGACGA--------UAUGGUAUGGGUUCAGC ..((..................((((.....))))(((.((((..((((((.(((((.((.----...----------.....))))))).))--------)).))..))))))).)) ( -20.80) >DroYak_CAF1 4263 110 - 1 UGGCACCUCUACGAAUGCACUACCAAUAGUCCGAGGAAUCCCACUCUUAUCUCCUUGGGUUGGGUUGGGUUCUACUUGGUUCUACUGCGUCCA--------CUCCUCAUGGGGUGAGC (((.(((.....((((.((...(((((...(((((((..............))))))))))))..)).)))).....))).)))....(..((--------((((....))))))..) ( -32.94) >consensus __UCACCCAUAUAAAUGUAUUACCAAUAACCCGAGGAAUCCCACUCUUAUCUUCUUGGGU_____UGG__________GUUCUACUCGCCCGACUCUC___CUCCUUAUGGGGUGAGC ......................(((...(((((((((..............))))))))).....)))................(((((((...................))))))). (-11.49 = -12.01 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:16 2006