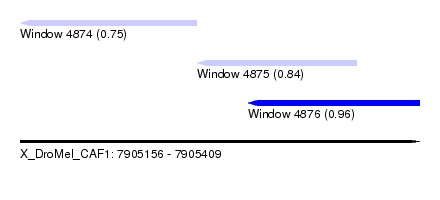
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,905,156 – 7,905,409 |
| Length | 253 |
| Max. P | 0.964758 |

| Location | 7,905,156 – 7,905,268 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -17.99 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7905156 112 - 22224390 GACAAAAUCAACAUCCAGCGGCAGCGCGGCCAUAAACAAAGCCAAAAUCAAAAGUCAUUCGGAUAUAUACAUAUAUUCGCCCAUAAUAGCGAAUAAACAGUAACAGAACGAG (((..............(((....)))(((..........)))..........)))..(((......(((...(((((((........)))))))....)))......))). ( -20.30) >DroSec_CAF1 5806 104 - 1 CACAAC------AUCCAGCGGCAGCGCGGCCAUAAACAAAGCCAAAAUCAAAAGUCAUUCGGAUAUAU--AUAUAUUUGCACAUAAAAGCGAAUAAACAGUAACAGAACGGG ......------((((.(((....)))(((..........))).................))))....--...(((((((........)))))))................. ( -17.30) >DroSim_CAF1 8925 110 - 1 CACAACAUCAACAUCCAGCGGCAGCGCGGCCAUAAACAAAGCCAAAAUCAAAAGUCAUUCGGAUAUAU--AUAUAUUUGCACAUAAAAGCGAAUAAACAGUAACAGAACGAG .......((...((((.(((....)))(((..........))).................))))....--...(((((((........)))))))..............)). ( -17.70) >consensus CACAACAUCAACAUCCAGCGGCAGCGCGGCCAUAAACAAAGCCAAAAUCAAAAGUCAUUCGGAUAUAU__AUAUAUUUGCACAUAAAAGCGAAUAAACAGUAACAGAACGAG .((.........((((.(((....)))(((..........))).................)))).........(((((((........)))))))....))........... (-17.99 = -17.77 + -0.22)

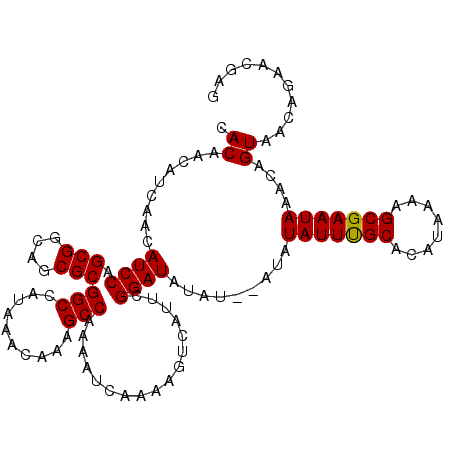

| Location | 7,905,268 – 7,905,369 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -14.07 |
| Consensus MFE | -13.23 |
| Energy contribution | -12.57 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7905268 101 - 22224390 AAUCUGUAUGUUGUAUAUGUUCAAUUCAAAAUUCAGCAACAAUAGCAACAACGCAAUUC--------AGCAACAACAACAAACAAAACCGACAAGCUAAACUUGGCUAA ....(((.((((((...((((..............((.......))......((.....--------.)))))))))))).))).........(((((....))))).. ( -15.00) >DroSec_CAF1 5910 107 - 1 AAUCUGUAUGUUGUAUAUGUUUAAUUUA-AAUUCAACAACAAAAGCAAAAACGCAAUUCAACAAUUCAGCAACAAC-ACAAGCAAAACCGACGAGCUAAACUCGGCUAA ....(((.((((((...((((.(((...-.))).))))......((......))..............))))))))-).............((((.....))))..... ( -13.60) >DroSim_CAF1 9035 107 - 1 AAUCUGUAUGUUGUAUAUGUUCAAUUUA-AAUUCAACAACAAAAGCAACAACGCAAUUCAACAAUUCAGCAACAAC-ACAAGCAAAACCGACGAGCUAAACUCGGCUAA ....(((.((((((...((((.(((...-.))).))))......((......))..............))))))))-).............((((.....))))..... ( -13.60) >consensus AAUCUGUAUGUUGUAUAUGUUCAAUUUA_AAUUCAACAACAAAAGCAACAACGCAAUUCAACAAUUCAGCAACAAC_ACAAGCAAAACCGACGAGCUAAACUCGGCUAA ....(((.((((((...((((.(((.....))).))))......((......))..............))))))...)))...........((((.....))))..... (-13.23 = -12.57 + -0.66)


| Location | 7,905,300 – 7,905,409 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -17.67 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7905300 109 - 22224390 UUUUUAUUUUUACAGUUGCAACUUUUUAACAAUCGUGUACAAUCUGUAUGUUGUAUAUGUUCAAUUCAAAAUUCAGCAACAAUAGCAACAACGCAAUUC--------AGCAACAACA ..............(((((..............((((((((((......)))))))))).........................((......)).....--------.))))).... ( -17.60) >DroSec_CAF1 5942 115 - 1 UUUUUAUUUUUACAGUUGCAACUUUUUAACAAUCGUGUACAAUCUGUAUGUUGUAUAUGUUUAAUUUA-AAUUCAACAACAAAAGCAAAAACGCAAUUCAACAAUUCAGCAACAAC- ..............(((((.....(((((....((((((((((......))))))))))......)))-)).............((......))..............)))))...- ( -17.70) >DroSim_CAF1 9067 115 - 1 UUUUUAUUUUUACAGUUGCAACUUUUUAACAAUCGUGUACAAUCUGUAUGUUGUAUAUGUUCAAUUUA-AAUUCAACAACAAAAGCAACAACGCAAUUCAACAAUUCAGCAACAAC- ..............(((((.....(((((....((((((((((......))))))))))......)))-)).............((......))..............)))))...- ( -17.70) >consensus UUUUUAUUUUUACAGUUGCAACUUUUUAACAAUCGUGUACAAUCUGUAUGUUGUAUAUGUUCAAUUUA_AAUUCAACAACAAAAGCAACAACGCAAUUCAACAAUUCAGCAACAAC_ ..............(((((..............((((((((((......)))))))))).........................((......))..............))))).... (-17.60 = -17.60 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:02 2006