
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,903,750 – 7,903,947 |
| Length | 197 |
| Max. P | 0.953878 |

| Location | 7,903,750 – 7,903,867 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -36.47 |
| Energy contribution | -36.92 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7903750 117 + 22224390 CUCGACUGAAAGUUGCAGCUGCUCAGGCGUGCGGUAGAGCGAGACAACAGUAGGGGGCUACGUAAUACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUU---UGCG (((.((((....((((.(((((........)))))...)))).....)))).)))(((((((.....)))))))....((((((((((((((.....)))))).....)))))---))). ( -45.10) >DroSec_CAF1 4355 120 + 1 CUCCGUUGAAAGUUGCAGCUGCUCAGGCGUGCGGUAGAGCGAGACAACAGUAGGGGGCUACGUAAUACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUUGUGUGCG ((((((........((....))........)))).)).(((..(((((((((...(((((((.....)))))))...)))..((((((((((.....)))).)))))).)))))).))). ( -46.99) >DroYak_CAF1 7582 112 + 1 CUCGGCUGAAAGUUGCAGCUGCUCAGGCGUGCGAUAGAACGAGACAACGGUAGGGGCCUACGUAA-ACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCG-CCAUGUGUAUU---U--- ((((..(....(((((((((.....))).)))))).)..))))......(((..(((.((((...-.)))))))...)))..((((((((((.....))))-)))))).....---.--- ( -42.40) >consensus CUCGGCUGAAAGUUGCAGCUGCUCAGGCGUGCGGUAGAGCGAGACAACAGUAGGGGGCUACGUAAUACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUU___UGCG ((((........((((((((.....))).))))).....))))......(((...(((((((.....)))))))...)))..((((((((((.....)))).))))))............ (-36.47 = -36.92 + 0.45)

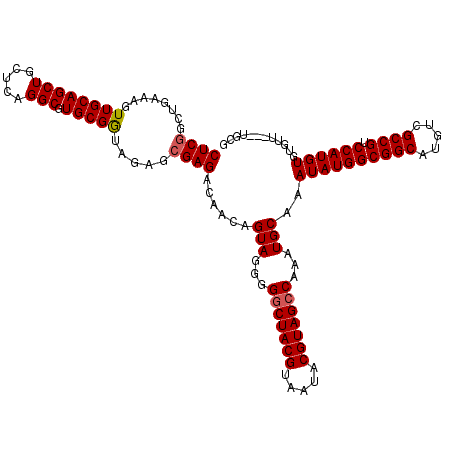

| Location | 7,903,790 – 7,903,907 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -43.21 |
| Consensus MFE | -32.84 |
| Energy contribution | -33.07 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7903790 117 + 22224390 GAGACAACAGUAGGGGGCUACGUAAUACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUU---UGCGUUCGAGUGUGUGUGUGUGAAGUGGCAUUCGGACGAGGAGA .(((((...(((...(((((((.....)))))))...)))..((((((((((.....)))).)))))).))))---).(((((((((((..............)))))))))))...... ( -46.14) >DroSec_CAF1 4395 117 + 1 GAGACAACAGUAGGGGGCUACGUAAUACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUUGUGUGCGGGUGCGUGUGUGUGUGCGAAGUGGCAUUCGGACG---AGA ...(((((((((...(((((((.....)))))))...)))..((((((((((.....)))).)))))).))))))((.(((((((...(((....))).....))))))).)).---... ( -48.20) >DroYak_CAF1 7622 107 + 1 GAGACAACGGUAGGGGCCUACGUAA-ACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCG-CCAUGUGUAUU---U--------GUGAGUGUGUGUGAAGUGGCAUUCGAACAGGGAGA ...((((..(((..(((.((((...-.)))))))...)))..((((((((((.....))))-))))))....)---)--------))((((((..........))))))........... ( -35.30) >consensus GAGACAACAGUAGGGGGCUACGUAAUACGUAGCCAAAUGCAAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUU___UGCG___G_GUGUGUGUGUGUGAAGUGGCAUUCGGACG_GGAGA ...(((((((((...(((((((.....)))))))...)))..((((((((((.....)))).))))))..................))).)))((.((((......)))).))....... (-32.84 = -33.07 + 0.23)

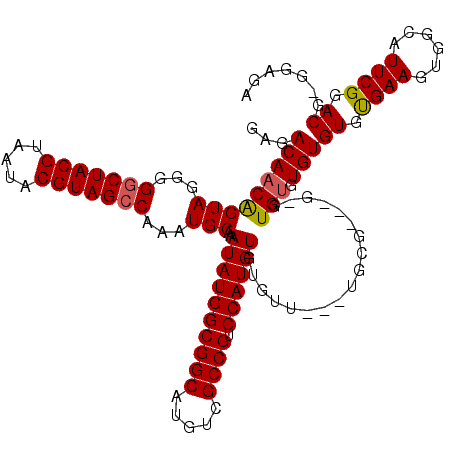

| Location | 7,903,790 – 7,903,907 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -22.65 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7903790 117 - 22224390 UCUCCUCGUCCGAAUGCCACUUCACACACACACACUCGAACGCA---AACACACAUGGACGGCGACAUGCCGCCAUAUUUGCAUUUGGCUACGUAUUACGUAGCCCCCUACUGUUGUCUC .......((.(((.((................)).))).))(((---((.....((((.((((.....)))))))).)))))....(((((((.....)))))))............... ( -28.79) >DroSec_CAF1 4395 117 - 1 UCU---CGUCCGAAUGCCACUUCGCACACACACACGCACCCGCACACAACACACAUGGACGGCGACAUGCCGCCAUAUUUGCAUUUGGCUACGUAUUACGUAGCCCCCUACUGUUGUCUC ...---.((.((((......)))).))........((....))..((((((...((((.((((.....))))))))..........(((((((.....)))))))......))))))... ( -32.90) >DroYak_CAF1 7622 107 - 1 UCUCCCUGUUCGAAUGCCACUUCACACACACUCAC--------A---AAUACACAUGG-CGGCGACAUGCCGCCAUAUUUGCAUUUGGCUACGU-UUACGUAGGCCCCUACCGUUGUCUC ...............(((................(--------(---(((.((.((((-((((.....))))))))...)).)))))..((((.-...)))))))............... ( -25.60) >consensus UCUCC_CGUCCGAAUGCCACUUCACACACACACAC_C___CGCA___AACACACAUGGACGGCGACAUGCCGCCAUAUUUGCAUUUGGCUACGUAUUACGUAGCCCCCUACUGUUGUCUC .........((((((((..................((....))...........((((.((((.....))))))))....))))))))(((((.....)))))................. (-22.65 = -22.93 + 0.28)


| Location | 7,903,830 – 7,903,947 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -33.37 |
| Energy contribution | -32.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7903830 117 + 22224390 AAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUU---UGCGUUCGAGUGUGUGUGUGUGAAGUGGCAUUCGGACGAGGAGAGCAUUUGUUGUUGUAGGUUUUAGUGCCAUCGUUGUGUAUC ..((((((((((.....)))).))))))(((((---(.(((((((((((..............)))))))))))....))))))...........(((......)))............. ( -35.24) >DroSec_CAF1 4435 117 + 1 AAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUUGUGUGCGGGUGCGUGUGUGUGUGCGAAGUGGCAUUCGGACG---AGAGCAUUUGUUGUUGUAGGUUUUAGUGCCAUCGUUGUGUAUC ..((((((((((.....)))).)))))).....(..((((....))))..)(((..(((.((((((((..(((.---(.((((.....)))).)..)))..))))))))..)))..))). ( -38.80) >DroYak_CAF1 7661 108 + 1 AAAUAUGGCGGCAUGUCGCCG-CCAUGUGUAUU---U--------GUGAGUGUGUGUGAAGUGGCAUUCGAACAGGGAGAGCAUUUGUUGUUGUAGGUUUUAGUGCCAUCGUUGUGUAUC ..((((((((((.....))))-)))))).....---.--------......(((..(((.((((((((.((((....(.((((.....)))).)..)))).))))))))..)))..))). ( -36.60) >consensus AAAUAUGGCGGCAUGUCGCCGUCCAUGUGUGUU___UGCG___G_GUGUGUGUGUGUGAAGUGGCAUUCGGACG_GGAGAGCAUUUGUUGUUGUAGGUUUUAGUGCCAUCGUUGUGUAUC ..((((((((((.....)))).)))))).......................(((..(((.((((((((..(((....(.((((.....)))).)..)))..))))))))..)))..))). (-33.37 = -32.93 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:55 2006