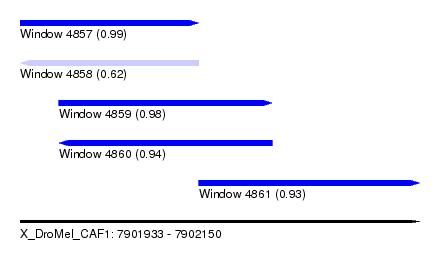
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,901,933 – 7,902,150 |
| Length | 217 |
| Max. P | 0.992357 |

| Location | 7,901,933 – 7,902,030 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -26.52 |
| Energy contribution | -26.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.68 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7901933 97 + 22224390 UCACAGAGAGCAACACCAAAAAAAAAAUAAAAAGAUGAGCCAGGAGCGGCGCUCUCAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACA ...................................((((((....(.(((((((((..((((......)))).....)))))))))))))))).... ( -27.50) >DroSec_CAF1 2550 91 + 1 UCACAGAGAGCAACACCAAAAAAA------AAAGAUGAGCCAGGAGCGGCGCUCUUAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACA ........................------.....((((((....(.(((((((((..((((......)))).....)))))))))))))))).... ( -25.70) >DroSim_CAF1 5664 91 + 1 UCACAGAGAGCAACACCAAAAAAA------AAAGAUGAGCCAGGAGCGGCGCUCUUAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACA ........................------.....((((((....(.(((((((((..((((......)))).....)))))))))))))))).... ( -25.70) >consensus UCACAGAGAGCAACACCAAAAAAA______AAAGAUGAGCCAGGAGCGGCGCUCUUAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACA ...................................((((((....(.(((((((((..((((......)))).....)))))))))))))))).... (-26.52 = -26.30 + -0.22)

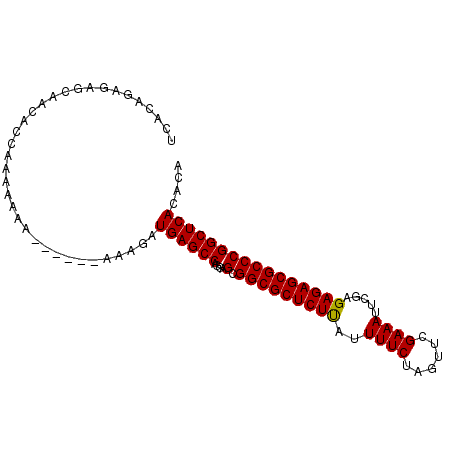

| Location | 7,901,933 – 7,902,030 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7901933 97 - 22224390 UGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUGAGAGCGCCGCUCCUGGCUCAUCUUUUUAUUUUUUUUUUGGUGUUGCUCUCUGUGA ...(((((((((((((((((.....((((......))))..))))))))).....)))))))).................................. ( -30.20) >DroSec_CAF1 2550 91 - 1 UGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUAAGAGCGCCGCUCCUGGCUCAUCUUU------UUUUUUUGGUGUUGCUCUCUGUGA ...((((((((((((((((....(.((((......)))).).)))))))).....))))))))....------........................ ( -26.20) >DroSim_CAF1 5664 91 - 1 UGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUAAGAGCGCCGCUCCUGGCUCAUCUUU------UUUUUUUGGUGUUGCUCUCUGUGA ...((((((((((((((((....(.((((......)))).).)))))))).....))))))))....------........................ ( -26.20) >consensus UGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUAAGAGCGCCGCUCCUGGCUCAUCUUU______UUUUUUUGGUGUUGCUCUCUGUGA ...((((((((((((((((......((((......))))...)))))))).....)))))))).................................. (-26.20 = -26.20 + -0.00)

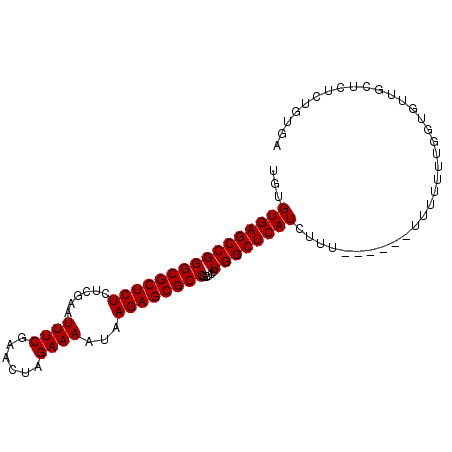
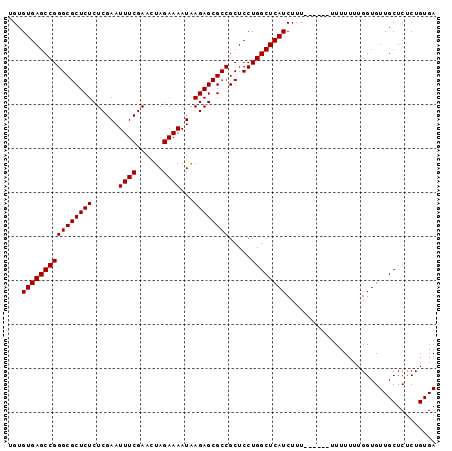
| Location | 7,901,954 – 7,902,070 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7901954 116 + 22224390 AAAAAAUAAAAAGAUGAGCCAG----GAGCGGCGCUCUCAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACAUACACGCAUACGCAGACCGCACAAUAGAGCUGUCUCGCUU ..............((((((..----..(.(((((((((..((((......)))).....))))))))))))))))...............((((((.((........)).)))).)).. ( -36.30) >DroSec_CAF1 2571 110 + 1 AAA------AAAGAUGAGCCAG----GAGCGGCGCUCUUAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACAUACACGCAUACGCAGACCGCACAAUAGAGCUGUCUCGCUU ...------.....((((((..----..(.(((((((((..((((......)))).....))))))))))))))))...............((((((.((........)).)))).)).. ( -34.50) >DroSim_CAF1 5685 110 + 1 AAA------AAAGAUGAGCCAG----GAGCGGCGCUCUUAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACAUACACGCAUAUGCAGACCGCACAAUAGAGCUGUCUCGCUU ...------.....((((((..----..(.(((((((((..((((......)))).....))))))))))))))))...............((((((.((........)).)))).)).. ( -34.20) >DroYak_CAF1 5745 114 + 1 AAA------AAAGAUGAGCCAGAGUGAAGCGGCGCUCUCAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACAUACACGCAUACGCAUACCGCACAAUAGAGCUGUCUCGCUU ...------.(((.((((.(((.((((.(((((((((((..((((......)))).....)))))))))..))))))........((....))................))).))))))) ( -34.70) >consensus AAA______AAAGAUGAGCCAG____GAGCGGCGCUCUCAUUUUCUAGUUCGAAAUUCGAGAGAGCGCCCGGCUCACACAUACACGCAUACGCAGACCGCACAAUAGAGCUGUCUCGCUU ..............((((((.(.......)(((((((((..((((......)))).....))))))))).))))))...............((((((.((........)).)))).)).. (-33.12 = -33.12 + 0.00)

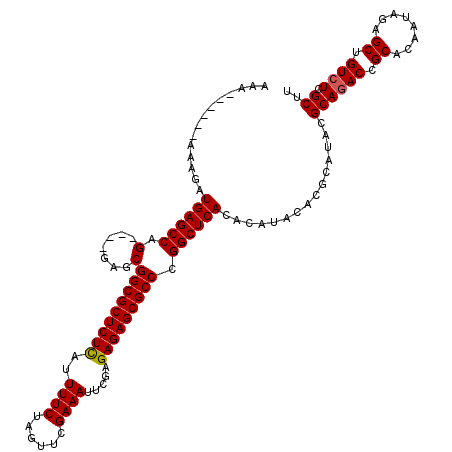

| Location | 7,901,954 – 7,902,070 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -36.66 |
| Energy contribution | -37.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7901954 116 - 22224390 AAGCGAGACAGCUCUAUUGUGCGGUCUGCGUAUGCGUGUAUGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUGAGAGCGCCGCUC----CUGGCUCAUCUUUUUAUUUUUU .....((((.((.(....).)).))))((((((....))))))(((((((((((((((((.....((((......))))..)))))))))....----.))))))))............. ( -40.70) >DroSec_CAF1 2571 110 - 1 AAGCGAGACAGCUCUAUUGUGCGGUCUGCGUAUGCGUGUAUGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUAAGAGCGCCGCUC----CUGGCUCAUCUUU------UUU .....((((.((.(....).)).))))((((((....))))))((((((((((((((((....(.((((......)))).).))))))))....----.))))))))....------... ( -36.70) >DroSim_CAF1 5685 110 - 1 AAGCGAGACAGCUCUAUUGUGCGGUCUGCAUAUGCGUGUAUGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUAAGAGCGCCGCUC----CUGGCUCAUCUUU------UUU .(((.((..(((.........((((..(((((((....)))))))..))))((((((((....(.((((......)))).).))))))))))).----)).))).......------... ( -38.30) >DroYak_CAF1 5745 114 - 1 AAGCGAGACAGCUCUAUUGUGCGGUAUGCGUAUGCGUGUAUGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUGAGAGCGCCGCUUCACUCUGGCUCAUCUUU------UUU .(((.(((..((.(....).)).(((((((....)))))))(((.((((..(((((((((.....((((......))))..))))))))))))))))))).))).......------... ( -39.90) >consensus AAGCGAGACAGCUCUAUUGUGCGGUCUGCGUAUGCGUGUAUGUGUGAGCCGGGCGCUCUCUCGAAUUUCGAACUAGAAAAUAAGAGCGCCGCUC____CUGGCUCAUCUUU______UUU .....((((.((.(....).)).))))((((((....))))))(((((((((((((((((.....((((......))))..))))))))).........))))))))............. (-36.66 = -37.23 + 0.56)


| Location | 7,902,030 – 7,902,150 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -29.29 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7902030 120 + 22224390 UACACGCAUACGCAGACCGCACAAUAGAGCUGUCUCGCUUGCACUUGAGGAAACAGAACAAGCAAAGCAGGCAAAUGGCGCACGCUAAACGCUGCAAAAUACGUUUAGUAAACCUUAAAA .....(((..((.((((.((........)).))))))..)))..((((((..((.((((.......((((.(...((((....))))...))))).......)))).))...)))))).. ( -29.34) >DroSec_CAF1 2641 120 + 1 UACACGCAUACGCAGACCGCACAAUAGAGCUGUCUCGCUUGCACUUGAGGAAACAGAGCAAGCAAAGCAGGCAAAUGGCGCACGCUAAACGCUGCAAAAUACGUUUUGUAAACCUUAAAA .....(((..((.((((.((........)).))))))..)))..((((((..(((((((.......((((.(...((((....))))...))))).......)))))))...)))))).. ( -33.14) >DroSim_CAF1 5755 120 + 1 UACACGCAUAUGCAGACCGCACAAUAGAGCUGUCUCGCUUGCACUUAAGGAAACAGAGCAAGCAAAGCAGGCAAAUGGCGCACGCUAAACGCUGCAAAAUACGUUUUGUAAACCUUAAAA .....(((...((((((.((........)).)))).)).)))..((((((..(((((((.......((((.(...((((....))))...))))).......)))))))...)))))).. ( -32.54) >DroYak_CAF1 5819 120 + 1 UACACGCAUACGCAUACCGCACAAUAGAGCUGUCUCGCUUGCACUUGAGGAUAAAGAGCAACCAAAGCAGGCAAAUGGCGCACGCUUAACGCUGCAAAAUAUGUUUUUUAAACCUAAAAA .....(((..((...((.((........)).))..))..))).....(((.(((((((((......((((.(....(((....)))....)))))......)))))))))..)))..... ( -23.60) >consensus UACACGCAUACGCAGACCGCACAAUAGAGCUGUCUCGCUUGCACUUGAGGAAACAGAGCAAGCAAAGCAGGCAAAUGGCGCACGCUAAACGCUGCAAAAUACGUUUUGUAAACCUUAAAA .....(((..((.((((.((........)).))))))..)))..((((((..(((((((.......((((.(...((((....))))...))))).......)))))))...)))))).. (-29.29 = -29.41 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:49 2006