
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,897,407 – 7,897,567 |
| Length | 160 |
| Max. P | 0.794478 |

| Location | 7,897,407 – 7,897,527 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -48.13 |
| Consensus MFE | -45.93 |
| Energy contribution | -46.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7897407 120 + 22224390 CUCCUGCUGCUGUCACACCGGCACCAAAGCCAGCACCGGCCGCGGCACCGCCGCCCGCCAUCUGGCUAAAGGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAA .......(((.........(((......))).((((.(((((((((...)))))..(((....)))....))))..(((((.((.....)).)))))))))...)))(((.....))).. ( -46.10) >DroSim_CAF1 1141 120 + 1 CUCCAGCUGCUGUCACACCGGCACCAAAGCCAGCACCGGCCGCGGCACCGCCGCCCGCCAGCUGGCUAAAGGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAA ...((((((...((((..((.......(((((((...(((.(((((...)))))..))).)))))))...(((....))).))..))))...)))))).........(((.....))).. ( -49.30) >DroYak_CAF1 1142 120 + 1 CUCCUGCUGCUGUCACACCGGCACCAAAGCCAGCACCGGCCGCGGCACCGCCGCCCGCCAGCUGGCUAAAUGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAA ....(((.(.((.(((...((((....(((((((...(((.(((((...)))))..))).)))))))...))))..(((((.((.....)).)))))))))).))))(((.....))).. ( -49.00) >consensus CUCCUGCUGCUGUCACACCGGCACCAAAGCCAGCACCGGCCGCGGCACCGCCGCCCGCCAGCUGGCUAAAGGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAA .....((.(.((.(((...(((.....(((((((...(((.(((((...)))))..))).)))))))....)))..(((((.((.....)).)))))))))).))).(((.....))).. (-45.93 = -46.27 + 0.33)


| Location | 7,897,407 – 7,897,527 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -60.70 |
| Consensus MFE | -57.33 |
| Energy contribution | -58.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7897407 120 - 22224390 UUCCCAAUGUGGGUGCGUUGCACAGCCAGGUUCACGACGUGGCCAUGGCCUUUAGCCAGAUGGCGGGCGGCGGUGCCGCGGCCGGUGCUGGCUUUGGUGCCGGUGUGACAGCAGCAGGAG ..(((.....)))(((((..(((.((((.(((...))).))))..((((((..((((((...((.(((.(((....))).))).)).))))))..)).)))))))..)).)))....... ( -58.70) >DroSim_CAF1 1141 120 - 1 UUCCCAAUGUGGGUGCGUUGCACAGCCAGGUUCACGACGUGGCCAUGGCCUUUAGCCAGCUGGCGGGCGGCGGUGCCGCGGCCGGUGCUGGCUUUGGUGCCGGUGUGACAGCAGCUGGAG .(((......((.(((((..(((.((((.(((...))).))))..((((((..(((((((..((.(((.(((....))).))).)))))))))..)).)))))))..)).))).))))). ( -61.50) >DroYak_CAF1 1142 120 - 1 UUCCCAAUGUGGGUGCGUUGCACAGCCAGGUUCACGACGUGGCCAUGGCAUUUAGCCAGCUGGCGGGCGGCGGUGCCGCGGCCGGUGCUGGCUUUGGUGCCGGUGUGACAGCAGCAGGAG ..(((.....)))(((((..(((.((((.(((...))).))))..(((((((.(((((((..((.(((.(((....))).))).)))))))))..))))))))))..)).)))....... ( -61.90) >consensus UUCCCAAUGUGGGUGCGUUGCACAGCCAGGUUCACGACGUGGCCAUGGCCUUUAGCCAGCUGGCGGGCGGCGGUGCCGCGGCCGGUGCUGGCUUUGGUGCCGGUGUGACAGCAGCAGGAG ..(((.....)))(((((..(((.((((.(((...))).))))..((((((..(((((((..((.(((.(((....))).))).)))))))))..)).)))))))..)).)))....... (-57.33 = -58.00 + 0.67)


| Location | 7,897,447 – 7,897,567 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -49.00 |
| Consensus MFE | -46.94 |
| Energy contribution | -46.83 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
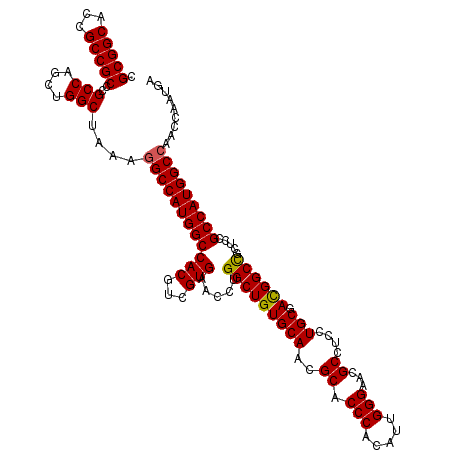
>X_DroMel_CAF1 7897447 120 + 22224390 CGCGGCACCGCCGCCCGCCAUCUGGCUAAAGGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAACGCCUCCUGCGACGGCCGCUCCGCCAUGGCCAACCAAUGA .(((((...)))))..(((....)))....((((((((((((...))).....(((((((((..((.(((.....)))...))....))).)))))).....)))))))))......... ( -50.40) >DroSim_CAF1 1181 120 + 1 CGCGGCACCGCCGCCCGCCAGCUGGCUAAAGGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAACGCCUCCUGCGACGGCCGCUCCGCCAUGGCCAACCAAUGA .(((((...)))))..(((....)))....((((((((((((...))).....(((((((((..((.(((.....)))...))....))).)))))).....)))))))))......... ( -50.40) >DroYak_CAF1 1182 120 + 1 CGCGGCACCGCCGCCCGCCAGCUGGCUAAAUGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAACGCCGCCUGCGAUGGCUGCUCCGCCAUGGCCAACCAACGA .(((((...)))))..((((..((((.....))))))))....((((.....((((((((....((.(((.....)))...((.(((......))).))...))))))))))....)))) ( -46.20) >consensus CGCGGCACCGCCGCCCGCCAGCUGGCUAAAGGCCAUGGCCACGUCGUGAACCUGGCUGUGCAACGCACCCACAUUGGGAACGCCUCCUGCGACGGCCGCUCCGCCAUGGCCAACCAAUGA .(((((...)))))..(((....)))....((((((((((((...))).....(((((((((..((.(((.....)))...))....))).)))))).....)))))))))......... (-46.94 = -46.83 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:34 2006