
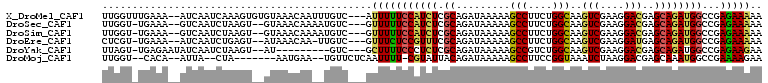
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,895,743 – 7,895,848 |
| Length | 105 |
| Max. P | 0.575683 |

| Location | 7,895,743 – 7,895,848 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.01 |
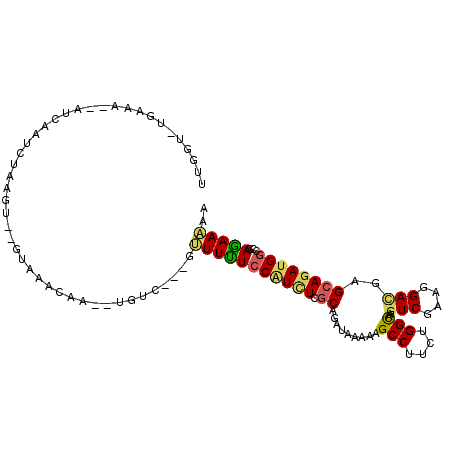
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7895743 105 - 22224390 UUGGUUUGAAA--AUCAAUCAAAGUGUGUAAACAAUUUGUC---AUUUUUCCAUCUCGCAGAUAAAAAGCCUUCUGGCAAGUCGAAGGACGAGCAGAUGGCCGAGAAAAA (((((..((((--(.....((((.(((....))).))))..---..)))))(((((.((.........(((....)))..(((....)))..))))))))))))...... ( -28.50) >DroSec_CAF1 6443 102 - 1 UUGGU-UGAAA--GUCAAUCUAAGU--GUAAACAAAAUGUC---GUUUUUCCAUCUCGCAGAUAAAAAGCCUUCUGGCAAGUCGAAGGACGAGCAGAUGGCCGAGAAAAA ..(((-((...--..))))).....--..........(.((---(.....((((((.((.........(((....)))..(((....)))..)))))))).))).).... ( -25.40) >DroSim_CAF1 6346 102 - 1 UUGGU-UGAAA--GUCAAUCUAAGU--GUAAACAAAAUGUC---GUUUUUCCAUCUCGCAGAUAAAAAGCCUUCUGGCAAGUCGAAGGACGAGCAGAUGGCCGAGAAAAA ..(((-((...--..))))).....--..........(.((---(.....((((((.((.........(((....)))..(((....)))..)))))))).))).).... ( -25.40) >DroEre_CAF1 6170 101 - 1 CUCGU-UGAAA--AUCAAUCUGAGU--AUAAACAA-UUGUC---GUUUCUCCGUUUCGCAGAUAAAAAGCCUUCUGGCAAGUCGAAGGAUGAGCAGAUGGCCGAGAAAAA ((((.-.((((--..((((.((...--.....)))-)))..---.)))).((((((.((.........(((....)))..(((....)))..)))))))).))))..... ( -22.60) >DroYak_CAF1 6357 95 - 1 UUAGU-UGAGAAUAUCAAUCUAAGU--AU---------GUC---GCUUUUCCCUCUCGCAGAUAAAAAGCCGUCUGGCAAGUCGAAGGACGAGCAGAUGGCCGAGAAGAA (((((-(((.....)))).))))..--..---------...---....(((..(((((..........((((((((.(..(((....)))..)))))))))))))).))) ( -27.30) >DroMoj_CAF1 12442 94 - 1 UUGGU--CACA--AUUA--CUA-------AAUGAA--UGUUCUCAAUUUU-CGUAUUACAGAUAAAAAGCCUUCCGGUAAAUCUAAGGACGAGCAAAUGGCCGAAAAGAA (((((--((..--....--...-------..(((.--.....)))...((-(((.....((((.....(((....)))..))))....)))))....)))))))...... ( -15.00) >consensus UUGGU_UGAAA__AUCAAUCUAAGU__GUAAACAA__UGUC___GUUUUUCCAUCUCGCAGAUAAAAAGCCUUCUGGCAAGUCGAAGGACGAGCAGAUGGCCGAGAAAAA .............................................(((((((((((.((.........(((....)))..(((....)))..))))))))...))))).. (-15.42 = -15.40 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:29 2006