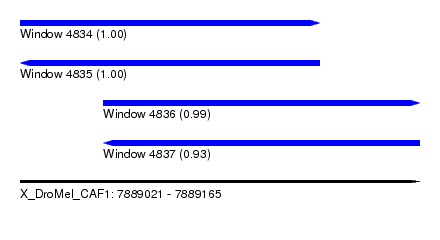
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,889,021 – 7,889,165 |
| Length | 144 |
| Max. P | 0.999987 |

| Location | 7,889,021 – 7,889,129 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.42 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -12.61 |
| Energy contribution | -14.99 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.48 |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7889021 108 + 22224390 --------ACACACUUUACACGAAUAUGUAUAUAUGGUUUACAGAGAUCUCUGGCCUAUGAUA--GCCAUAACAUAAGAUUUGGUAAUCACAAAUCUCUGUAAACAUAUACACCGACU --------..................(((((((...((((((((((((...((((........--))))........((((....))))....)))))))))))))))))))...... ( -26.60) >DroSec_CAF1 2909 109 + 1 --------ACACACUUCACACGAAUAUGUAUAUAUGGUUCACAGAGAUCGAUGGCCUACAAUAUGGCUAU-ACAUAAGAUUUGGUGAUCACAAAUCUCUGUGAACAUAUACACCGACU --------..................(((((((...((((((((((((..((((((........))))))-......((((....))))....)))))))))))))))))))...... ( -32.80) >DroEre_CAF1 2940 83 + 1 --------ACACACUUCACACGAAUC-GGGCGCAUGGUUCACAGAAACCUCUGGACUA--------------------------UAAUAACAAAUCUCUGUGACCAUAUACACCGACU --------................((-((....(((((.((((((.............--------------------------............))))))))))).....)))).. ( -17.11) >DroYak_CAF1 3008 100 + 1 AAAGAAAAACACAUUUUACACAAAUC-GGGCGUAUGGUGCACAGAAAUCGCUGGAAUA-------UCC-----ACCA-----GUUAAUAACAAAUCUCUGUGCCCAUAAACGCCGACU ..........................-.(((((((((.(((((((....(((((....-------...-----.)))-----))............)))))))))))..))))).... ( -27.59) >consensus ________ACACACUUCACACGAAUA_GGACAUAUGGUUCACAGAAAUCGCUGGACUA_______GCC_____AUAA_____GGUAAUAACAAAUCUCUGUGAACAUAUACACCGACU ...........................((((((((((.((((((((((...((.....................................)).))))))))))))))))))))..... (-12.61 = -14.99 + 2.38)


| Location | 7,889,021 – 7,889,129 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.42 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -21.02 |
| Energy contribution | -22.15 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.65 |
| SVM decision value | 5.47 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7889021 108 - 22224390 AGUCGGUGUAUAUGUUUACAGAGAUUUGUGAUUACCAAAUCUUAUGUUAUGGC--UAUCAUAGGCCAGAGAUCUCUGUAAACCAUAUAUACAUAUUCGUGUAAAGUGUGU-------- .....((((((((((((((((((((((..((((....))))........((((--(......))))).))))))))))))).)))))))))...................-------- ( -34.10) >DroSec_CAF1 2909 109 - 1 AGUCGGUGUAUAUGUUCACAGAGAUUUGUGAUCACCAAAUCUUAUGU-AUAGCCAUAUUGUAGGCCAUCGAUCUCUGUGAACCAUAUAUACAUAUUCGUGUGAAGUGUGU-------- .....(((((((((((((((((((((.(((..(..(((....((((.-.....)))))))..)..))).)))))))))))).)))))))))...................-------- ( -33.90) >DroEre_CAF1 2940 83 - 1 AGUCGGUGUAUAUGGUCACAGAGAUUUGUUAUUA--------------------------UAGUCCAGAGGUUUCUGUGAACCAUGCGCCC-GAUUCGUGUGAAGUGUGU-------- ((((((.(..(((((((((((((((((.(.....--------------------------......).))))))))))).))))))..)))-))))..............-------- ( -28.00) >DroYak_CAF1 3008 100 - 1 AGUCGGCGUUUAUGGGCACAGAGAUUUGUUAUUAAC-----UGGU-----GGA-------UAUUCCAGCGAUUUCUGUGCACCAUACGCCC-GAUUUGUGUAAAAUGUGUUUUUCUUU ((((((.((.((((((((((((((((.((.....))-----.(.(-----((.-------....))).)))))))))))).))))).))))-))))...................... ( -33.90) >consensus AGUCGGUGUAUAUGGUCACAGAGAUUUGUGAUUACC_____UUAU_____GGC_______UAGGCCAGAGAUCUCUGUGAACCAUACACAC_GAUUCGUGUAAAGUGUGU________ ((((.((((((((((((((((((((((.((...................................)).)))))))))))).)))))))))).))))...................... (-21.02 = -22.15 + 1.13)



| Location | 7,889,051 – 7,889,165 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -16.35 |
| Energy contribution | -16.66 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7889051 114 + 22224390 UUACAGAGAUCUCUGGCCUAUGAUA--GCCAUAACAUAAGAUUUGGUAAUCACAAAUCUCUGUAAACAUAUACACCGACUAUGCUCUACUUACACAUUUGUGAUGUGGGCGUGUUG ((((((((((...((((........--))))........((((....))))....))))))))))..........((((.((((.(((((((((....))))).)))))))))))) ( -32.10) >DroSec_CAF1 2939 115 + 1 UCACAGAGAUCGAUGGCCUACAAUAUGGCUAU-ACAUAAGAUUUGGUGAUCACAAAUCUCUGUGAACAUAUACACCGACUAUGCUCUACUUACACAUUUGUGAUGUGGGCGUGUUG ((((((((((..((((((........))))))-......((((....))))....))))))))))..........((((.((((.(((((((((....))))).)))))))))))) ( -38.30) >DroEre_CAF1 2969 90 + 1 UCACAGAAACCUCUGGACUA--------------------------UAAUAACAAAUCUCUGUGACCAUAUACACCGACUAUGUUGUACUUACACAUUUGUGAUGUGGGCGUGUUA (((((((.............--------------------------............)))))))((((((.(((.....((((.((....))))))..)))))))))........ ( -17.01) >DroYak_CAF1 3045 99 + 1 GCACAGAAAUCGCUGGAAUA-------UCC-----ACCA-----GUUAAUAACAAAUCUCUGUGCCCAUAAACGCCGACUAUGUGGUACUUACACAUUUGCGAUGUGGGCGUAUUG (((((((....(((((....-------...-----.)))-----))............)))))))......(((((.((.(((((.......))))).......)).))))).... ( -27.59) >consensus UCACAGAAAUCGCUGGACUA_______GCC_____AUAA_____GGUAAUAACAAAUCUCUGUGAACAUAUACACCGACUAUGCUCUACUUACACAUUUGUGAUGUGGGCGUGUUG ((((((((((...((.....................................)).))))))))))..............(((((.(((((((((....))))).)))))))))... (-16.35 = -16.66 + 0.31)

| Location | 7,889,051 – 7,889,165 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -20.44 |
| Energy contribution | -19.88 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7889051 114 - 22224390 CAACACGCCCACAUCACAAAUGUGUAAGUAGAGCAUAGUCGGUGUAUAUGUUUACAGAGAUUUGUGAUUACCAAAUCUUAUGUUAUGGC--UAUCAUAGGCCAGAGAUCUCUGUAA ....((((((((((.....)))))......((......)))))))......((((((((((((..((((....))))........((((--(......))))).)))))))))))) ( -29.80) >DroSec_CAF1 2939 115 - 1 CAACACGCCCACAUCACAAAUGUGUAAGUAGAGCAUAGUCGGUGUAUAUGUUCACAGAGAUUUGUGAUCACCAAAUCUUAUGU-AUAGCCAUAUUGUAGGCCAUCGAUCUCUGUGA ....((((((((((.....)))))......((......)))))))......(((((((((((.(((..(..(((....((((.-.....)))))))..)..))).))))))))))) ( -29.60) >DroEre_CAF1 2969 90 - 1 UAACACGCCCACAUCACAAAUGUGUAAGUACAACAUAGUCGGUGUAUAUGGUCACAGAGAUUUGUUAUUA--------------------------UAGUCCAGAGGUUUCUGUGA ........(((.(((((...(((((.......)))))....))).)).)))((((((((((((.(.....--------------------------......).)))))))))))) ( -20.00) >DroYak_CAF1 3045 99 - 1 CAAUACGCCCACAUCGCAAAUGUGUAAGUACCACAUAGUCGGCGUUUAUGGGCACAGAGAUUUGUUAUUAAC-----UGGU-----GGA-------UAUUCCAGCGAUUUCUGUGC ......(((((...(((...(((((.......)))))....)))....)))))(((((((((.((.....))-----.(.(-----((.-------....))).)))))))))).. ( -28.70) >consensus CAACACGCCCACAUCACAAAUGUGUAAGUACAACAUAGUCGGUGUAUAUGGUCACAGAGAUUUGUGAUUACC_____UUAU_____GGC_______UAGGCCAGAGAUCUCUGUGA ....(((((.((..(((....)))...(.....)...)).)))))......((((((((((((.((...................................)).)))))))))))) (-20.44 = -19.88 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:27 2006