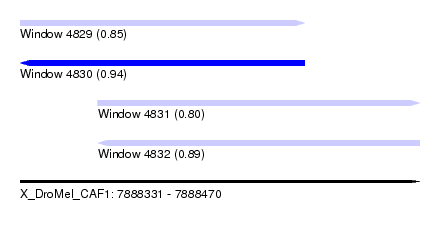
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,888,331 – 7,888,470 |
| Length | 139 |
| Max. P | 0.944383 |

| Location | 7,888,331 – 7,888,430 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -42.18 |
| Consensus MFE | -27.97 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
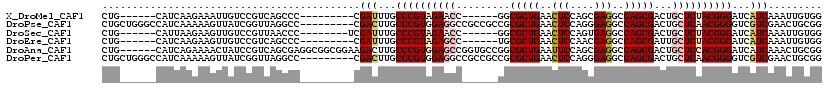
>X_DroMel_CAF1 7888331 99 + 22224390 CUG------CAUCAAGAAAUUGUCCGUCAGCCC---------CGAUUUGCCCGUAGAACC------GGCGCUGAACUCCAGCGAGGCCAGCGACUGCUCUACGGGAUCAUCAAAUUGUGG (((------(..(((....)))...).))).((---------(((((((((((((((..(------(((((((..(((....)))..))))).))).)))))))).....))))))).)) ( -37.60) >DroPse_CAF1 1972 111 + 1 CUGCUGGGCCAUCAAAAAGUUAUCGGUUAGGCC---------CGACUUGCCCGUGGAGGCCGCCGCCGCGCUGAACUCCAGGGAGGCCAGCGACUGCUCAACGGGGUCGUCGAACUGCGG ((((.((((((((...........)))..))))---------)(((...(((((.(((((....))).(((((..(((....)))..))))).....)).)))))...))).....)))) ( -43.70) >DroSec_CAF1 2219 100 + 1 CUG------CAUUAAGAAGUUGUCCGUUAACCC--------UCGAUUUGCCCGUAGAACC------GGCGCUGAACUCCAGUGAGGCCAGCGACUGCUCUACGGGAUCAUCAAAUUGUGG ...------.........((((.....))))((--------.(((((((((((((((..(------(((((((..(((....)))..))))).))).)))))))).....))))))).)) ( -35.80) >DroEre_CAF1 2244 99 + 1 CUG------CAUCAAGAAGUUGUCCGUCAGCCC---------CGAUUUGCCCGUAGAGCC------UGCGCUGAACUCCAACGAGGCCAGCGAUUGCUCUACGGGAUCAUCAAAUUGUGG ...------.........((((.....))))((---------(((((((((((((((((.------..(((((..(((....)))..)))))...)))))))))).....))))))).)) ( -41.10) >DroAna_CAF1 2107 114 + 1 CUG------CAUCAGAAAACUAUCCGUCAGCGAGGCGGCGGAAGACUUGCCCGUGGAGCCGGUGCCGGCGCUGAAUUCCAGCGAGGCCAGCGACUGCUCCACGGGAUCAUCAAACUGCGG (((------((...........((((((.((...)))))))).((....((((((((((((.((((..(((((.....))))).)).)).))...))))))))))....))....))))) ( -51.20) >DroPer_CAF1 7123 111 + 1 CUGCUGGGCCAUCAAAAAGUUAUCGGUUAGGCC---------CGACUUGCCCGUGGAGGCCGCCGCCGCGCUGAACUCCAGGGAGGCCAGCGACUGCUCAACGGGGUCGUCGAACUGCGG ((((.((((((((...........)))..))))---------)(((...(((((.(((((....))).(((((..(((....)))..))))).....)).)))))...))).....)))) ( -43.70) >consensus CUG______CAUCAAAAAGUUAUCCGUCAGCCC_________CGACUUGCCCGUAGAGCC______GGCGCUGAACUCCAGCGAGGCCAGCGACUGCUCUACGGGAUCAUCAAACUGCGG ...........................................(((...((((((((((.........(((((..(((....)))..)))))...))))))))))...)))......... (-27.97 = -28.50 + 0.53)


| Location | 7,888,331 – 7,888,430 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7888331 99 - 22224390 CCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUGGCCUCGCUGGAGUUCAGCGCC------GGUUCUACGGGCAAAUCG---------GGGCUGACGGACAAUUUCUUGAUG------CAG ......((((((...((((((((((.(.(((((..(((....)))..))))).)------.))))))))))...))))---------))((....(((.....)))....)------).. ( -38.10) >DroPse_CAF1 1972 111 - 1 CCGCAGUUCGACGACCCCGUUGAGCAGUCGCUGGCCUCCCUGGAGUUCAGCGCGGCGGCGGCCUCCACGGGCAAGUCG---------GGCCUAACCGAUAACUUUUUGAUGGCCCAGCAG ((((.((((((((....)))))))).(.(((((..(((....)))..)))))).))))..((((....))))..((.(---------((((....(((.......)))..))))).)).. ( -49.20) >DroSec_CAF1 2219 100 - 1 CCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUGGCCUCACUGGAGUUCAGCGCC------GGUUCUACGGGCAAAUCGA--------GGGUUAACGGACAACUUCUUAAUG------CAG .......(((((...((((((((((.(.(((((..(((....)))..))))).)------.))))))))))...)))))--------(.(((((.(((....)))))))).------).. ( -39.30) >DroEre_CAF1 2244 99 - 1 CCACAAUUUGAUGAUCCCGUAGAGCAAUCGCUGGCCUCGUUGGAGUUCAGCGCA------GGCUCUACGGGCAAAUCG---------GGGCUGACGGACAACUUCUUGAUG------CAG ......((((((...((((((((((...(((((..(((....)))..)))))..------.))))))))))...))))---------))((....(((.....)))....)------).. ( -40.40) >DroAna_CAF1 2107 114 - 1 CCGCAGUUUGAUGAUCCCGUGGAGCAGUCGCUGGCCUCGCUGGAAUUCAGCGCCGGCACCGGCUCCACGGGCAAGUCUUCCGCCGCCUCGCUGACGGAUAGUUUUCUGAUG------CAG ..(((....(((...((((((((((.(..((((((...((((.....))))))))))..).))))))))))...))).(((((.((...)).).))))...........))------).. ( -49.40) >DroPer_CAF1 7123 111 - 1 CCGCAGUUCGACGACCCCGUUGAGCAGUCGCUGGCCUCCCUGGAGUUCAGCGCGGCGGCGGCCUCCACGGGCAAGUCG---------GGCCUAACCGAUAACUUUUUGAUGGCCCAGCAG ((((.((((((((....)))))))).(.(((((..(((....)))..)))))).))))..((((....))))..((.(---------((((....(((.......)))..))))).)).. ( -49.20) >consensus CCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUGGCCUCGCUGGAGUUCAGCGCC______GGCUCCACGGGCAAAUCG_________GGGCUAACGGACAACUUCUUGAUG______CAG .........(((...((((((((((...(((((..(((....)))..))))).........))))))))))...)))........................................... (-30.03 = -30.00 + -0.03)



| Location | 7,888,358 – 7,888,470 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -32.36 |
| Energy contribution | -33.14 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7888358 112 + 22224390 --CGAUUUGCCCGUAGAACC------GGCGCUGAACUCCAGCGAGGCCAGCGACUGCUCUACGGGAUCAUCAAAUUGUGGUAUCUGUUUGAGGCCUGAGGCCAGCGAGGCCAGUGAGGCA --.(((...((((((((..(------(((((((..(((....)))..))))).))).))))))))))).((((((.(......).)))))).((((.(((((.....))))..).)))). ( -46.30) >DroSec_CAF1 2246 105 + 1 -UCGAUUUGCCCGUAGAACC------GGCGCUGAACUCCAGUGAGGCCAGCGACUGCUCUACGGGAUCAUCAAAUUGUGGUAUCUGUUUGAGGCCCGAGGCCAGC--------CGAGGCA -..(((...((((((((..(------(((((((..(((....)))..))))).))).))))))))...)))...(((.(((.((.....)).)))))).(((...--------...))). ( -41.70) >DroEre_CAF1 2271 103 + 1 --CGAUUUGCCCGUAGAGCC------UGCGCUGAACUCCAACGAGGCCAGCGAUUGCUCUACGGGAUCAUCAAAUUGUGGUAUCUGUUUGAGGCCCGAGGCCAG---------UGAGGCA --.(((...((((((((((.------..(((((..(((....)))..)))))...))))))))))...)))...(((.(((.((.....)).)))))).(((..---------...))). ( -46.10) >DroYak_CAF1 2311 103 + 1 --CGAUUUGCCCGUAGAGCC------GGCGCUGAACUCCAACGAGGCAAGCGACUGCUCUACGGGAUCAUCAAAUUGUGGUAUCUGUUUGAGGCCCGAGGCCAG---------CGAGGCA --.(((...((((((((((.------((((((...(((....)))...)))).))))))))))))...)))...(((.(((.((.....)).)))))).(((..---------...))). ( -42.90) >DroAna_CAF1 2141 109 + 1 GAAGACUUGCCCGUGGAGCCGGUGCCGGCGCUGAAUUCCAGCGAGGCCAGCGACUGCUCCACGGGAUCAUCAAACUGCGGAAUCUGUUUCAGGCCGGCGGCCAG---------CGAAU-- ((.((....((((((((((((.((((..(((((.....))))).)).)).))...)))))))))).)).))....(((((((....)))).((((...)))).)---------))...-- ( -47.10) >DroPer_CAF1 7156 109 + 1 --CGACUUGCCCGUGGAGGCCGCCGCCGCGCUGAACUCCAGGGAGGCCAGCGACUGCUCAACGGGGUCGUCGAACUGCGGGAUUUGCUUCAGACCCGAUGACAG---------CGAGGCA --...(((((((((.(((((....))).(((((..(((....)))..))))).....)).)))).(((((((..(((.((......)).)))...))))))).)---------))))... ( -43.60) >consensus __CGAUUUGCCCGUAGAGCC______GGCGCUGAACUCCAGCGAGGCCAGCGACUGCUCUACGGGAUCAUCAAAUUGUGGUAUCUGUUUGAGGCCCGAGGCCAG_________CGAGGCA ...(((...((((((((((.......(((((((..(((....)))..))))).))))))))))))...)))....................(((.....))).................. (-32.36 = -33.14 + 0.78)


| Location | 7,888,358 – 7,888,470 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -44.05 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7888358 112 - 22224390 UGCCUCACUGGCCUCGCUGGCCUCAGGCCUCAAACAGAUACCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUGGCCUCGCUGGAGUUCAGCGCC------GGUUCUACGGGCAAAUCG-- .((((....((((.....))))..)))).....................(((...((((((((((.(.(((((..(((....)))..))))).)------.))))))))))...))).-- ( -47.90) >DroSec_CAF1 2246 105 - 1 UGCCUCG--------GCUGGCCUCGGGCCUCAAACAGAUACCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUGGCCUCACUGGAGUUCAGCGCC------GGUUCUACGGGCAAAUCGA- .((....--------)).((((...))))..................(((((...((((((((((.(.(((((..(((....)))..))))).)------.))))))))))...)))))- ( -42.80) >DroEre_CAF1 2271 103 - 1 UGCCUCA---------CUGGCCUCGGGCCUCAAACAGAUACCACAAUUUGAUGAUCCCGUAGAGCAAUCGCUGGCCUCGUUGGAGUUCAGCGCA------GGCUCUACGGGCAAAUCG-- .(((...---------..)))....((..((.....))..)).......(((...((((((((((...(((((..(((....)))..)))))..------.))))))))))...))).-- ( -42.80) >DroYak_CAF1 2311 103 - 1 UGCCUCG---------CUGGCCUCGGGCCUCAAACAGAUACCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUUGCCUCGUUGGAGUUCAGCGCC------GGCUCUACGGGCAAAUCG-- .(((...---------..)))....((..((.....))..)).......(((...((((((((((.(.((((...(((....)))...)))).)------.))))))))))...))).-- ( -36.60) >DroAna_CAF1 2141 109 - 1 --AUUCG---------CUGGCCGCCGGCCUGAAACAGAUUCCGCAGUUUGAUGAUCCCGUGGAGCAGUCGCUGGCCUCGCUGGAAUUCAGCGCCGGCACCGGCUCCACGGGCAAGUCUUC --....(---------(.((((...)))).(((.....))).)).....(((...((((((((((.(..((((((...((((.....))))))))))..).))))))))))...)))... ( -47.20) >DroPer_CAF1 7156 109 - 1 UGCCUCG---------CUGUCAUCGGGUCUGAAGCAAAUCCCGCAGUUCGACGACCCCGUUGAGCAGUCGCUGGCCUCCCUGGAGUUCAGCGCGGCGGCGGCCUCCACGGGCAAGUCG-- .....((---------(((((...(((..(......)..)))((.((((((((....))))))))....((((..(((....)))..)))))))))))))((((....))))......-- ( -47.00) >consensus UGCCUCG_________CUGGCCUCGGGCCUCAAACAGAUACCACAAUUUGAUGAUCCCGUAGAGCAGUCGCUGGCCUCGCUGGAGUUCAGCGCC______GGCUCUACGGGCAAAUCG__ ..................(((.....)))....................(((...((((((((((...((((((.(((....))).)))))).........))))))))))...)))... (-33.72 = -33.75 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:22 2006