
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,887,591 – 7,887,705 |
| Length | 114 |
| Max. P | 0.970931 |

| Location | 7,887,591 – 7,887,705 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -52.00 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7887591 114 + 22224390 GCCUGCGCUGAUGCAACAGCA---GCAGCCGCCGCCGCAUUUGGAUGAAGCUUCAUGUCCGAGGGCGACUGCUGCGGCGGCAACACCUUCUGAUGCUGCUGCAAUUGCUGCUUGUCU ......((....((((..(((---(((((.((((((((.(((((((..........)))))))(((....)))))))))))..((.....))..))))))))..)))).))...... ( -48.20) >DroSec_CAF1 1488 111 + 1 CCCUGCGCUGAUGCA---GCA---GCAGCCGCCGCCGCGUUCGGAUGAAGCUUCAUGUCCGAGGGCGACUGCUGCGGCGGCAACACCUUCUGGUGCUGCUGCAAUUGCUGCUUGUCC ....((((...((((---(((---((.(((((((((((.(((((((..........))))))).)))......))))))))...(((....))))))))))))...)).))...... ( -57.40) >DroEre_CAF1 1537 108 + 1 CCCUGCGCUGACGC------A---GCCGCCGCCGCCGCGCUCGAAUGAAGCUUCAUGUCCGAGGGCGACUGCUGCGGCGGCAACACCUUUUGAUGAUGCUGCAAUUGCUGCUUGUCC ...((((....)))------)---((((((((.(((((.((((.((((....))))...)))).)))...)).))))))))..........((..(.((.((....)).)))..)). ( -46.10) >DroYak_CAF1 1562 117 + 1 CCCUGCGCAGAUGCUGCAGCAGCCGCCGCCGCCGCCGCGCUCGUAUGAAGCUUCAUGUCCGAGGGCGACUGCUGUGGCGGCAACACCUUUUGAUGCUGCUGCAAUUGCUGCUUGUCC ......((((....(((((((((.((((((((.(((((.(((((((((....)))))..)))).)))...)).))))))))..((.....))..)))))))))....))))...... ( -56.30) >consensus CCCUGCGCUGAUGCA___GCA___GCAGCCGCCGCCGCGCUCGGAUGAAGCUUCAUGUCCGAGGGCGACUGCUGCGGCGGCAACACCUUCUGAUGCUGCUGCAAUUGCUGCUUGUCC ....(((....)))..........(((((.((.(.(((.(((((((..........))))))).))).).))((((((((((.((.....)).))))))))))...)))))...... (-37.06 = -37.50 + 0.44)

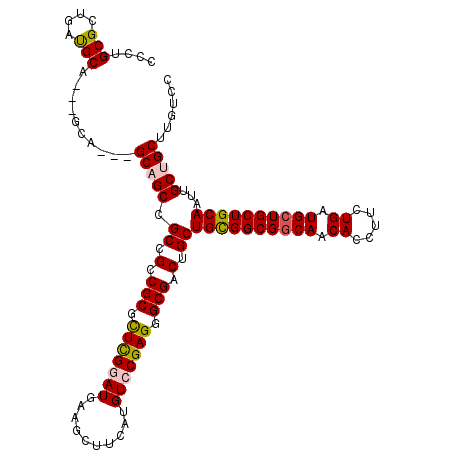

| Location | 7,887,591 – 7,887,705 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -53.88 |
| Consensus MFE | -41.45 |
| Energy contribution | -42.45 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7887591 114 - 22224390 AGACAAGCAGCAAUUGCAGCAGCAUCAGAAGGUGUUGCCGCCGCAGCAGUCGCCCUCGGACAUGAAGCUUCAUCCAAAUGCGGCGGCGGCUGC---UGCUGUUGCAUCAGCGCAGGC ......((((((...((.((((((((....)))))))).)).((((((((((((.(((....))).(((.(((....))).))))))))))))---)))))))))............ ( -51.60) >DroSec_CAF1 1488 111 - 1 GGACAAGCAGCAAUUGCAGCAGCACCAGAAGGUGUUGCCGCCGCAGCAGUCGCCCUCGGACAUGAAGCUUCAUCCGAACGCGGCGGCGGCUGC---UGC---UGCAUCAGCGCAGGG ......((.((...((((((((((((....)))(((((((((((.((....))..(((((..(((....))))))))..))))))))))).))---)))---))))...)))).... ( -56.20) >DroEre_CAF1 1537 108 - 1 GGACAAGCAGCAAUUGCAGCAUCAUCAAAAGGUGUUGCCGCCGCAGCAGUCGCCCUCGGACAUGAAGCUUCAUUCGAGCGCGGCGGCGGCGGC---U------GCGUCAGCGCAGGG .(((..(((((....((((((((.......))))))))((((((....(((((.((((((..(((....))))))))).))))).))))))))---)------)))))......... ( -51.40) >DroYak_CAF1 1562 117 - 1 GGACAAGCAGCAAUUGCAGCAGCAUCAAAAGGUGUUGCCGCCACAGCAGUCGCCCUCGGACAUGAAGCUUCAUACGAGCGCGGCGGCGGCGGCGGCUGCUGCAGCAUCUGCGCAGGG ......((((...(((((((((((((....))))))((((((.(.((.(((((.((((...((((....)))).)))).))))).)).).)))))).)))))))...))))...... ( -56.30) >consensus GGACAAGCAGCAAUUGCAGCAGCAUCAAAAGGUGUUGCCGCCGCAGCAGUCGCCCUCGGACAUGAAGCUUCAUCCGAACGCGGCGGCGGCGGC___UGC___UGCAUCAGCGCAGGG ......((.((...(((.((((((((....)))))))).(((((....(((((.((((((..(((....))))))))).))))).))))).............)))...)))).... (-41.45 = -42.45 + 1.00)

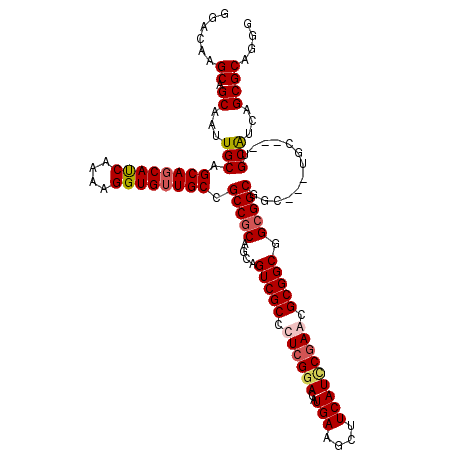
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:19 2006