
| Sequence ID | X_DroMel_CAF1 |
|---|---|
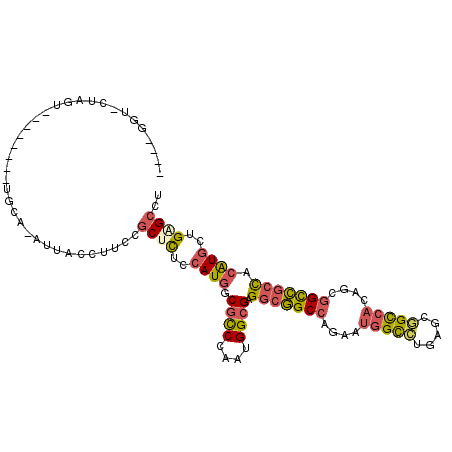
| Location | 7,886,568 – 7,886,665 |
| Length | 97 |
| Max. P | 0.982535 |

| Location | 7,886,568 – 7,886,665 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
| Mean single sequence MFE | -43.98 |
| Consensus MFE | -31.03 |
| Energy contribution | -31.45 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7886568 97 + 22224390 ----GGU-CUAGUU-------UGGA-AUUACCUUCCGCUCUCCAUGGCGCCUAAUGGCGAGGCGGCCAGAAUGGCCUGAGCGGCCACAGCGGCCGCCACAUGCUGAGCCU ----...-......-------.(((-(.....))))((((..((((.((((....)))).(((((((....(((((.....)))))....))))))).))))..)))).. ( -46.90) >DroPse_CAF1 406 98 + 1 ----GGCAUCAACG-------AGCA-CUUACCUGCCACUCUCCAUAGCCUCCAGGGGCGAGGAGGCCAGGACGGUCUGGGCUGUCAUUGUAGUCGAUACGUUCUGGGCCU ----(((......(-------.(((-......))))..(((((...((((....))))..)))))((((((((...(.(((((......))))).)..))))))))))). ( -37.10) >DroSec_CAF1 473 104 + 1 ----GGU-CCGGUUUUAGGAUUGGG-AUUACCUUCCGCUCUCCAUGGCGCCCAAUGGCGAGGCGGCCAGAAUGGCCUGAGCGGCCACAGCGGCCGCCACAUGCUGAGCCU ----(((-((.......))))).((-(......)))((((..((((.((((....)))).(((((((....(((((.....)))))....))))))).))))..)))).. ( -49.90) >DroEre_CAF1 525 101 + 1 ----GGU-CUUGU---GGGACUGCG-AUUACCUUCCGCUCUCCAUGGCGCCCAAGGGCGAGGCGGCCAGAAUGGCCUGAGCGGCCACAGCGGCCGCCACAUGCUGCGCCU ----(((-.(..(---(((...(((-.........)))..))))..).)))...(((((.(((((((.....))))((.((((((.....))))))))...))).))))) ( -46.50) >DroYak_CAF1 537 105 + 1 UGGUCGU-CUGGU---GGGAUUGUA-AUUACCUUCCGCUCUCCAUGGCGCCCAAUGGCGAGGCGGCCAAAAUAGCCUGUGCGGCAACAGCGGCCGCCACAUGUUGGGCCU ..(((((-..(((---((((..((.-...)).)))))))....)))))((((((((..(.(((((((......(((.....)))......))))))).).)))))))).. ( -42.60) >DroAna_CAF1 351 96 + 1 ----GGCAAUAG----------GUAACAUACCUUCCGCUUUCCAUGGCGCCCAAAGGCGAGGCUGCCAGUAUGGCUUGGGCAGCCACAGCGGCGGCCACAUGCUGCGCCU ----(((...((----------(((...)))))...((....((((.((((....)))).(((((((.((.(((((.....)))))..))))))))).))))..))))). ( -40.90) >consensus ____GGU_CUAGU________UGCA_AUUACCUUCCGCUCUCCAUGGCGCCCAAUGGCGAGGCGGCCAGAAUGGCCUGAGCGGCCACAGCGGCCGCCACAUGCUGAGCCU ....................................((((..((((.((((....)))).(((((((....(((((.....)))))....))))))).))))..)))).. (-31.03 = -31.45 + 0.42)


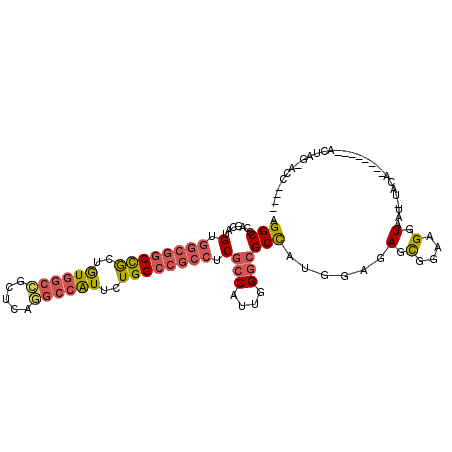
| Location | 7,886,568 – 7,886,665 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.81 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -31.62 |
| Energy contribution | -32.40 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7886568 97 - 22224390 AGGCUCAGCAUGUGGCGGCCGCUGUGGCCGCUCAGGCCAUUCUGGCCGCCUCGCCAUUAGGCGCCAUGGAGAGCGGAAGGUAAU-UCCA-------AACUAG-ACC---- ..((((..((((.((((((((..((((((.....))))))..)))))))).((((....)))).))))..))))((((.....)-))).-------......-...---- ( -50.00) >DroPse_CAF1 406 98 - 1 AGGCCCAGAACGUAUCGACUACAAUGACAGCCCAGACCGUCCUGGCCUCCUCGCCCCUGGAGGCUAUGGAGAGUGGCAGGUAAG-UGCU-------CGUUGAUGCC---- ...........((((((((..((...((.(((...((..(((((((((((........)))))))).)))..)))))..))...-))..-------.)))))))).---- ( -34.20) >DroSec_CAF1 473 104 - 1 AGGCUCAGCAUGUGGCGGCCGCUGUGGCCGCUCAGGCCAUUCUGGCCGCCUCGCCAUUGGGCGCCAUGGAGAGCGGAAGGUAAU-CCCAAUCCUAAAACCGG-ACC---- ..((((..((((.((((((((..((((((.....))))))..)))))))).((((....)))).))))..))))(((......)-))...(((.......))-)..---- ( -49.00) >DroEre_CAF1 525 101 - 1 AGGCGCAGCAUGUGGCGGCCGCUGUGGCCGCUCAGGCCAUUCUGGCCGCCUCGCCCUUGGGCGCCAUGGAGAGCGGAAGGUAAU-CGCAGUCCC---ACAAG-ACC---- .(((.......(.((((((((..((((((.....))))))..)))))))).)(((....))))))..(((..((((.......)-)))..))).---.....-...---- ( -49.40) >DroYak_CAF1 537 105 - 1 AGGCCCAACAUGUGGCGGCCGCUGUUGCCGCACAGGCUAUUUUGGCCGCCUCGCCAUUGGGCGCCAUGGAGAGCGGAAGGUAAU-UACAAUCCC---ACCAG-ACGACCA ..((((((..((.((((((((..((.(((.....))).))..)))))))).))...))))))....(((...(.(((..((...-.))..))))---.))).-....... ( -39.80) >DroAna_CAF1 351 96 - 1 AGGCGCAGCAUGUGGCCGCCGCUGUGGCUGCCCAAGCCAUACUGGCAGCCUCGCCUUUGGGCGCCAUGGAAAGCGGAAGGUAUGUUAC----------CUAUUGCC---- .(((.......(.(((.((((.(((((((.....))))))).)))).))).)(((....)))))).......((((.(((((...)))----------)).)))).---- ( -42.00) >consensus AGGCCCAGCAUGUGGCGGCCGCUGUGGCCGCUCAGGCCAUUCUGGCCGCCUCGCCAUUGGGCGCCAUGGAGAGCGGAAGGUAAU_UACA________ACUAG_ACC____ .(((.......(.((((((((..((((((.....))))))..)))))))).)(((....))))))......(.(....).)............................. (-31.62 = -32.40 + 0.78)

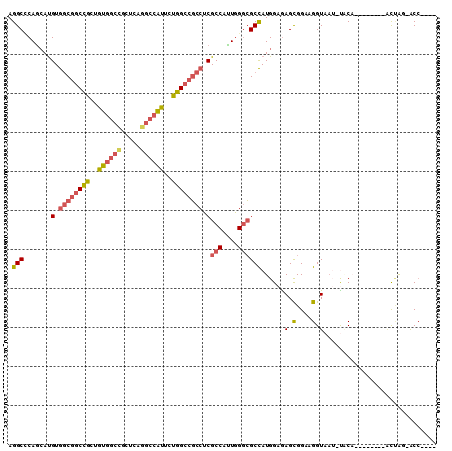
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:15 2006