
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,853,351 – 7,853,528 |
| Length | 177 |
| Max. P | 0.639829 |

| Location | 7,853,351 – 7,853,469 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7853351 118 - 22224390 AAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCUCAAGCAGAAGUGGAAGCCGAAGCGAAAUAACAAAGCAAAUAAACAAACAAAACGGCCAGGGGAAAGGACUGAAG ........((((((((....))))))))((((((((.....((....))....)))))...))).((...........))................(((((........)).)))... ( -24.40) >DroEre_CAF1 38132 118 - 1 AAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCUCAAGCAGAAGUGGAAGCCGAAGCGAAAUAACAAAGCAAAUAAACAAACAAAACGGGCAAAGGAAAGGACUUAAG .......(((((((((....))))))))).((((.......((....))..........(((...((...........))...........(.....))))........))))..... ( -24.70) >DroAna_CAF1 22915 112 - 1 AAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCCCGAGCAAAAGUGGAAGCCGAGGCGGAAUAACAAAGCAAAUAAACAAACAACAAGGGAAG------UGGCCAAAA .......(((((((((....))))))))).(.((((.....(((((.((..........)))).)))(......)...........................)------))).).... ( -25.80) >consensus AAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCUCAAGCAGAAGUGGAAGCCGAAGCGAAAUAACAAAGCAAAUAAACAAACAAAACGGGCAG_GGAAAGGACUAAAG ........((((((((....))))))))((((((((.....((....))....)))))...))).((...........))...................................... (-20.20 = -20.20 + -0.00)



| Location | 7,853,389 – 7,853,509 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -25.37 |
| Energy contribution | -25.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7853389 120 - 22224390 CAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCUCAAGCAGAAGUGGAAGCCGAAGCGAAAUAACAAAGC ..((..(((.((((..((.......(((((......)))))......(((((((((....))))))))).))..))))...((....)).....)))..))................... ( -25.60) >DroYak_CAF1 33683 120 - 1 CAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCUCAAGCAGAAGUGGAAGCCGAAGCGAAAUAACAAAGC ..((..(((.((((..((.......(((((......)))))......(((((((((....))))))))).))..))))...((....)).....)))..))................... ( -25.60) >DroAna_CAF1 22947 120 - 1 CAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCCCGAGCAAAAGUGGAAGCCGAGGCGGAAUAACAAAGC ......((..((((..((.......(((((......)))))......(((((((((....))))))))).))..))))...(((((.((..........)))).)))))........... ( -29.30) >consensus CAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAAAGCUCAAGCAGAAGUGGAAGCCGAAGCGAAAUAACAAAGC ..((..(((.((((..((.......(((((......)))))......(((((((((....))))))))).))..))))...((....)).....)))..))................... (-25.37 = -25.37 + -0.00)


| Location | 7,853,429 – 7,853,528 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7853429 99 + 22224390 UUAUGCGGACGAUUGGCAACAAAAGCUGCCAAUUUGUAUUUCAAUUAAAUUUGAAUGUAUUUGUGGGCACUUGGCUGCUGCUUUCUCC-------CCAGCUUUUCC-------------- ...((.(((.((..((((.((.(((.((((.((..(((((((((......))))).))))..)).)))))))...)).)))).)))))-------.))........-------------- ( -23.80) >DroYak_CAF1 33723 100 + 1 UUAUGCGGACGAUUGGCAACAAAAGCUGCCAAUUUGUAUUUCAAUUAAAUUUGAAUGUAUUUGUGGGCACUUGGCUGCUGCUUUUCCC-------CCAGCUUUUCCC------------- ....((((..((..((((.((.(((.((((.((..(((((((((......))))).))))..)).)))))))...)).))))..))..-------)).)).......------------- ( -24.10) >DroAna_CAF1 22987 120 + 1 UUAUGCGGACGAUUGGCAACAAAAGCUGCCAAUUUGUAUUUCAAUUAAAUUUGAAUGUAUUUGUGGGCACUUGGCUGCUGCUUUUCUCUUUUUCUUUAUCUUUUUCCCUCGGCCUUCGUC .......(((((..(((.....(((.((((.((..(((((((((......))))).))))..)).)))))))(((....))).............................))).))))) ( -25.70) >consensus UUAUGCGGACGAUUGGCAACAAAAGCUGCCAAUUUGUAUUUCAAUUAAAUUUGAAUGUAUUUGUGGGCACUUGGCUGCUGCUUUUCCC_______CCAGCUUUUCCC_____________ ....((((.((.(((....)))(((.((((.((..(((((((((......))))).))))..)).)))))))...))))))....................................... (-20.60 = -20.60 + -0.00)


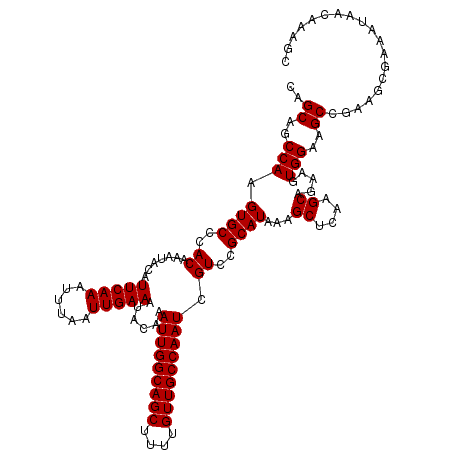
| Location | 7,853,429 – 7,853,528 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7853429 99 - 22224390 --------------GGAAAAGCUGG-------GGAGAAAGCAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAA --------------((....((((.-------........))))..))..((((..((.......(((((......)))))......(((((((((....))))))))).))..)))).. ( -24.60) >DroYak_CAF1 33723 100 - 1 -------------GGGAAAAGCUGG-------GGGAAAAGCAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAA -------------.((....((((.-------........))))..))..((((..((.......(((((......)))))......(((((((((....))))))))).))..)))).. ( -24.60) >DroAna_CAF1 22987 120 - 1 GACGAAGGCCGAGGGAAAAAGAUAAAGAAAAAGAGAAAAGCAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAA (((((.((((...((...............................))..).)))..........(((((......))))).......((((((((....)))))))))))))....... ( -23.28) >consensus _____________GGGAAAAGCUGG_______GGGAAAAGCAGCAGCCAAGUGCCCACAAAUACAUUCAAAUUUAAUUGAAAUACAAAUUGGCAGCUUUUGUUGCCAAUCGUCCGCAUAA .......................................((....))...((((..((.......(((((......)))))......(((((((((....))))))))).))..)))).. (-19.20 = -19.20 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:08 2006