
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,850,856 – 7,851,084 |
| Length | 228 |
| Max. P | 0.998178 |

| Location | 7,850,856 – 7,850,974 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850856 118 + 22224390 UAUAUGGUGUAUGCUAUAUAUGCUAUAUAUGCA-AUACAUAUAGGAUAUGGUACGUGGAUGGCACAGGUCAGGGGGUCAGGAGAGGCCAAUGAUGGCCAUUCUGCGACGUUUGCCAAUU ....((((((((.((((((.(((.......)))-....)))))).)))).((.((..((((((....((((...((((......))))..)))).)))).))..))))....))))... ( -36.70) >DroEre_CAF1 35616 104 + 1 --CUU--------AUAUAUA-----CAUACAUAUAUAUGGUCUACAUAUAGGAUAUGGAUGGCACAGGUUAGGGGGUCAGGAGGGGCCAAUGAUGGCCAUUCUGCGACGUUUGCCAAUU --...--------.......-----(((((...((((((.....)))))).).))))..(((((.(.(((.....(.(((((..(((((....))))).))))))))).).)))))... ( -29.20) >DroYak_CAF1 31145 111 + 1 UACUU--------AUAUAUAUACUACAAACUUAUAUAUAUAUAGAAUAUGGUACGUGGAUGGCACAGGUCAGGGGGUCAGGAGAGGCCAAUGAUGGCCAUUCUGCGACGUUUGCCAAUU ..(((--------(((((((((.........)))))))))).))....(((((((..((((((....((((...((((......))))..)))).)))).))..)).....)))))... ( -31.40) >consensus UACUU________AUAUAUAU_CUACAUACAUAUAUAUAUAUAGAAUAUGGUACGUGGAUGGCACAGGUCAGGGGGUCAGGAGAGGCCAAUGAUGGCCAUUCUGCGACGUUUGCCAAUU ..............((((((((.........))))))))....................(((((.(.(((.....(.(((((..(((((....))))).))))))))).).)))))... (-23.13 = -23.80 + 0.67)


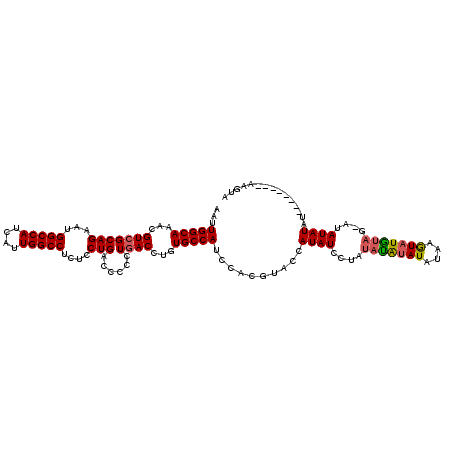
| Location | 7,850,856 – 7,850,974 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850856 118 - 22224390 AAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCUCUCCUGACCCCCUGACCUGUGCCAUCCACGUACCAUAUCCUAUAUGUAU-UGCAUAUAUAGCAUAUAUAGCAUACACCAUAUA ...(((((...(((((((...(((((....)))))....)))......))))...))))).....((...((((.((((((((..-...)))))))).))))...))............ ( -28.00) >DroEre_CAF1 35616 104 - 1 AAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCCCUCCUGACCCCCUAACCUGUGCCAUCCAUAUCCUAUAUGUAGACCAUAUAUAUGUAUG-----UAUAUAU--------AAG-- ...(((((...((((..((..(((((....)))))..)).))))...........))))).........((((((((...(((((....)))))-----)))))))--------)..-- ( -22.84) >DroYak_CAF1 31145 111 - 1 AAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCUCUCCUGACCCCCUGACCUGUGCCAUCCACGUACCAUAUUCUAUAUAUAUAUAAGUUUGUAGUAUAUAUAU--------AAGUA ...(((((...(((((((...(((((....)))))....)))......))))...)))))................(((((((((((.........))))))))))--------).... ( -27.30) >consensus AAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCUCUCCUGACCCCCUGACCUGUGCCAUCCACGUACCAUAUCCUAUAUAUAUAUAAGUAUGUAG_AUAUAUAU________AAGUA ...(((((...(((((((...(((((....)))))....)))......))))...)))))..........((((....(((((((....)))))))....))))............... (-20.52 = -20.97 + 0.45)


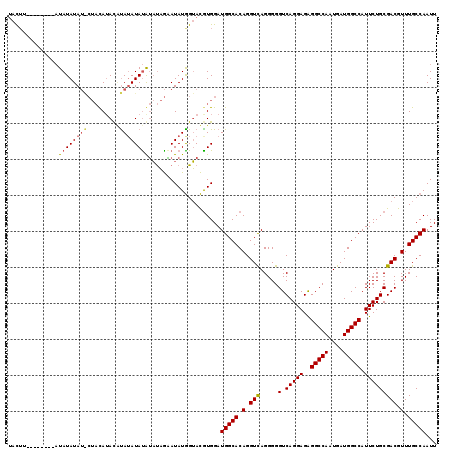
| Location | 7,850,894 – 7,851,014 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -25.47 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850894 120 - 22224390 AUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGAAAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCUCUCCUGACCCCCUGACCUGUGCCAUCCACGUACCAUAUCCUAUA ..((((((((((.(((...))).))))))..............(((((...(((((((...(((((....)))))....)))......))))...))))).......))))......... ( -27.60) >DroEre_CAF1 35640 120 - 1 AUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGAAAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCCCUCCUGACCCCCUAACCUGUGCCAUCCAUAUCCUAUAUGUAGAC .(((((((((((.(((...))).))))).)))))).(((....(((((...((((..((..(((((....)))))..)).))))...........)))))..(((((...)))))))).. ( -24.54) >DroYak_CAF1 31176 120 - 1 AUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGAAAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCUCUCCUGACCCCCUGACCUGUGCCAUCCACGUACCAUAUUCUAUA ..((((((((((.(((...))).))))))..............(((((...(((((((...(((((....)))))....)))......))))...))))).......))))......... ( -27.60) >consensus AUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGAAAUUGGCAAACGUCGCAGAAUGGCCAUCAUUGGCCUCUCCUGACCCCCUGACCUGUGCCAUCCACGUACCAUAUCCUAUA .(((((((((((.(((...))).))))).))))))........(((((...(((((((...(((((....)))))....)))......))))...))))).................... (-25.47 = -25.80 + 0.33)

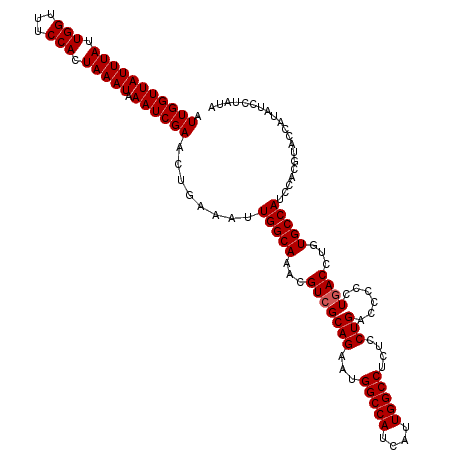
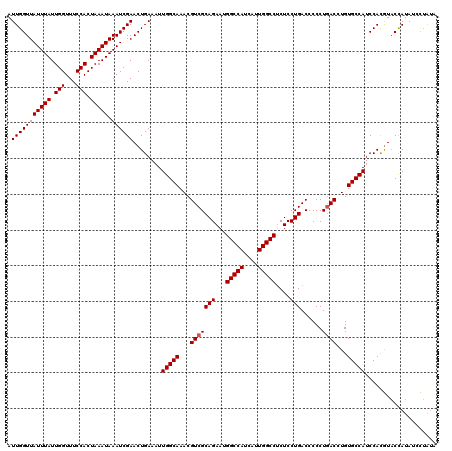
| Location | 7,850,974 – 7,851,084 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.89 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850974 110 - 22224390 GCACCACUGUGCACACACUGCACUCGGGGGAACGAGUUGUGCAAGUGUU----------UGUGCGAGCAGGACAUACAUCAUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGA ((((((((.((((((......(((((......)))))))))))))))..----------.))))...(((((......)).(((((((((((.(((...))).))))).)))))).))). ( -30.60) >DroEre_CAF1 35720 106 - 1 GCCCCACUGUGCA--CACUGCACUCGGGGGACAGAGUUGUGCAAGUGU------------UUGCGAGCAGGACAUACAUCAUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGA .((((...((((.--....))))...)))).(((.(((.(((..((..------------..))..))).)))........(((((((((((.(((...))).))))).)))))).))). ( -33.10) >DroYak_CAF1 31256 120 - 1 GCACCACUGUGCACACACUGCACUCGGGGGACAGAGUUGUGCAAGUGUGUGUGUGUGUGUGUGCGAGCAGGACAUACAUCAUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGA (((((((((..((((((((((((((.(....).))...)))).))))))))..)).))).))))...(((((......)).(((((((((((.(((...))).))))).)))))).))). ( -42.10) >DroAna_CAF1 21361 104 - 1 GUUCCACUGUCCA--UACUGCACUCAUGGGACAGAGUUCCGCAAGUGUG--------------UGAGCGAGACAUACAUCAUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGA ((..(.((((((.--.............)))))).(((((((....)))--------------.)))))..))........(((((((((((.(((...))).))))).))))))..... ( -24.34) >consensus GCACCACUGUGCA__CACUGCACUCGGGGGACAGAGUUGUGCAAGUGUG___________GUGCGAGCAGGACAUACAUCAUUGGUUAUUUAUUGGUUUCCACUAAAUAAAUCGAACUGA .((((...(((((.....)))))...)))).(((.(((.(((..((................))..))).)))........(((((((((((.(((...))).))))).)))))).))). (-20.95 = -21.89 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:04 2006