
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,850,602 – 7,850,738 |
| Length | 136 |
| Max. P | 0.989929 |

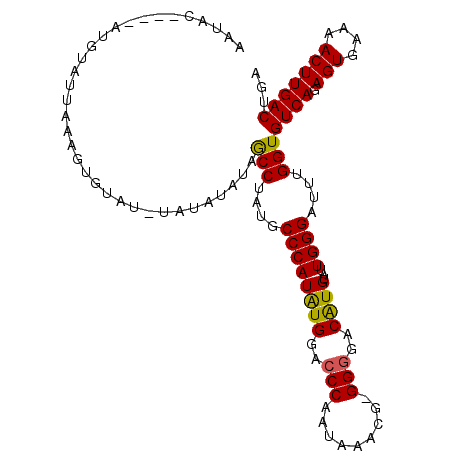
| Location | 7,850,602 – 7,850,699 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -19.96 |
| Energy contribution | -21.13 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850602 97 + 22224390 AAUAC------GUAUUAAAGUC----UAUAUAUAGCCUAUGCCCAUAUGGACCCAAUAAACG-GGGGACAUGUAUUGGGAUUUGGUGUCAGAGUGAAAACUUGACUGA .....------.......((((----........(((....((((((((..(((........-)))..))))...))))....)))....((((....)))))))).. ( -26.70) >DroEre_CAF1 35372 107 + 1 AAUACUCGUAUGUAUUAAAGUGUAUAUGUAUAUAGCCUAUGCCCAUAUGGACCCAAUAAACG-GGAGACGUGUAUUGGGAUCUGGUGUCAGAGUGAAAACUUGACUGA .(((..(((((((((....)))))))))..))).(((.....((....)).(((((((.(((-.....))).)))))))....)))((((.(((....)))))))... ( -28.00) >DroYak_CAF1 30898 99 + 1 ---------AUGUAUAUAAGUGUAUUUAUAUAUGACCCAUGCCCAUGUGGACCCAAUAAAAGAGGGGACAUGUAUUGGGAUUUGGUGUCAGAGUGAAAACUUGACUGA ---------..(((((((((....)))))))))((((((..((((.(((..(((.........)))..)))....))))...))).))).((((....))))...... ( -30.00) >consensus AAUAC____AUGUAUUAAAGUGUAU_UAUAUAUAGCCUAUGCCCAUAUGGACCCAAUAAACG_GGGGACAUGUAUUGGGAUUUGGUGUCAGAGUGAAAACUUGACUGA ..................................(((....((((((((..(((.........)))..))))...))))....)))((((.(((....)))))))... (-19.96 = -21.13 + 1.17)


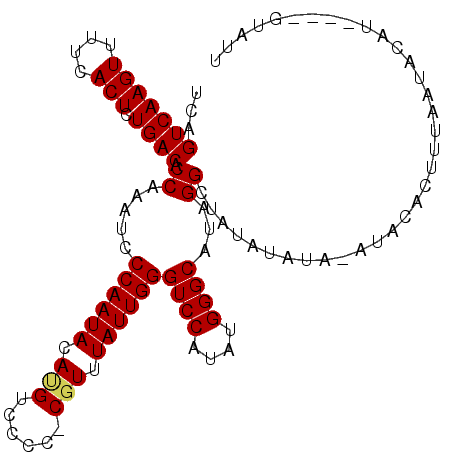
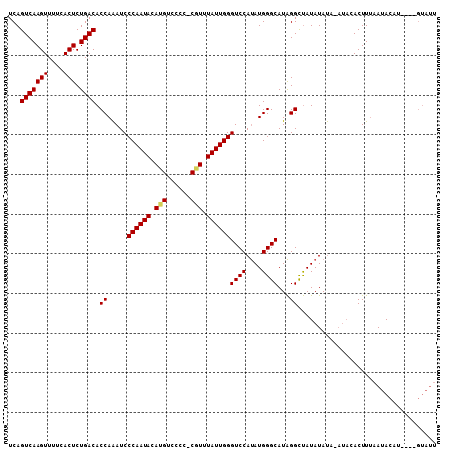
| Location | 7,850,602 – 7,850,699 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850602 97 - 22224390 UCAGUCAAGUUUUCACUCUGACACCAAAUCCCAAUACAUGUCCCC-CGUUUAUUGGGUCCAUAUGGGCAUAGGCUAUAUAUA----GACUUUAAUAC------GUAUU ...(((((((....))).))))........((((((.(((.....-))).))))))(((.((((((((....))).))))).----)))........------..... ( -18.50) >DroEre_CAF1 35372 107 - 1 UCAGUCAAGUUUUCACUCUGACACCAGAUCCCAAUACACGUCUCC-CGUUUAUUGGGUCCAUAUGGGCAUAGGCUAUAUACAUAUACACUUUAAUACAUACGAGUAUU ...(((((((....))).))))....((.(((((((.(((.....-))).))))))))).((((((.(...).)))))).............(((((......))))) ( -21.40) >DroYak_CAF1 30898 99 - 1 UCAGUCAAGUUUUCACUCUGACACCAAAUCCCAAUACAUGUCCCCUCUUUUAUUGGGUCCACAUGGGCAUGGGUCAUAUAUAAAUACACUUAUAUACAU--------- ...(((((((....))).)))).......((((...(((((.(((.........)))...)))))....))))...(((((((......)))))))...--------- ( -20.30) >consensus UCAGUCAAGUUUUCACUCUGACACCAAAUCCCAAUACAUGUCCCC_CGUUUAUUGGGUCCAUAUGGGCAUAGGCUAUAUAUA_AUACACUUUAAUACAU____GUAUU ...(((((((....))).)))).((.....((((((.(((......))).))))))((((....))))...))................................... (-17.61 = -17.83 + 0.22)

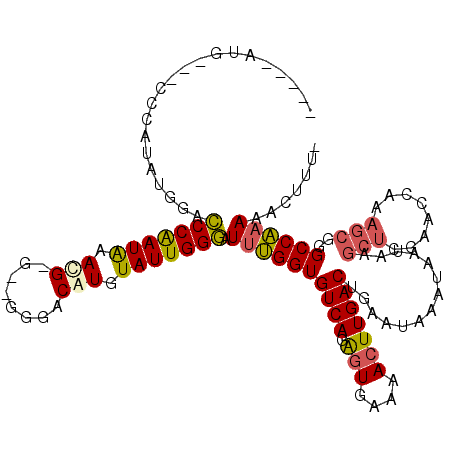
| Location | 7,850,630 – 7,850,738 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -18.38 |
| Energy contribution | -20.25 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7850630 108 + 22224390 -----AUG---CCCAUAUGGACCCAAUAAACG-G--GGGACAUGUAUUGGGAUUUGGUGUCAGAGUGAAAACUUGACUGAAUAAAUAAUAAGCUCGAACCAAAGCGGGCCAAAAACUUU- -----..(---(((.......(((((((....-(--....)...))))))).((((((((((.(((....)))))))(((.............))).))))))..))))..........- ( -30.82) >DroEre_CAF1 35410 108 + 1 -----AUG---CCCAUAUGGACCCAAUAAACG-G--GAGACGUGUAUUGGGAUCUGGUGUCAGAGUGAAAACUUGACUGAAUAAAUAAUAAGCUCGAACCAAAGCGGGCCAAAAACUUU- -----..(---(((....((((((((((.(((-.--....))).))))))).)))...((((.(((....)))))))..............(((........)))))))..........- ( -31.40) >DroYak_CAF1 30927 109 + 1 -----AUG---CCCAUGUGGACCCAAUAAAAGAG--GGGACAUGUAUUGGGAUUUGGUGUCAGAGUGAAAACUUGACUGAAUAAAUAAUAAGCUGGAACCAAAGCGGGCCAAAAACUUU- -----..(---(((..(((..(((.........)--))..)))((.((((..((..((((((.(((....)))))))..............))..)).)))).))))))..........- ( -30.33) >DroAna_CAF1 21038 110 + 1 AAAUGAAGCCACAUAUAGGGAUCCGAUGGAUG-UAUUGGACGUGUAUUGGAUUUCGGUGUCAGGCUGAAAACUUGACUGAAUAAAUAAUAAAC---------GACGAGCCGAAAAGUUUU ......((((((((...(..((((((((.(((-(.....)))).))))))))..).))))..)))).(((((((...................---------...........))))))) ( -27.51) >consensus _____AUG___CCCAUAUGGACCCAAUAAACG_G__GGGACAUGUAUUGGGAUUUGGUGUCAGAGUGAAAACUUGACUGAAUAAAUAAUAAGCUCGAACCAAAGCGGGCCAAAAACUUU_ .....................(((((((.(((........))).))))))).((((((((((.(((....)))))))..............(((........)))..))))))....... (-18.38 = -20.25 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:01 2006