
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,834,828 – 7,834,926 |
| Length | 98 |
| Max. P | 0.999444 |

| Location | 7,834,828 – 7,834,926 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.05 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -10.63 |
| Energy contribution | -11.75 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
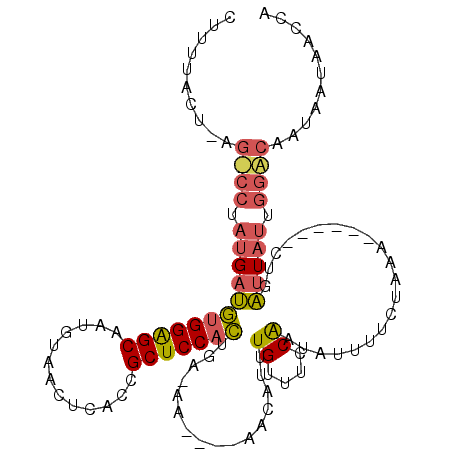
>X_DroMel_CAF1 7834828 98 + 22224390 CUUUUACUCAGCCCUAUGAUGUGGAGCAAUAUACCUCACCGCUCCACU---------AACAUUUGUUUCCAAUGUUUUCUAAG------CUGAUUUUUGGGCAAUAAUAGCCA .......(((((........(((((((.............))))))).---------((((((.......))))))......)------))))......(((.......))). ( -23.22) >DroSim_CAF1 16073 101 + 1 CUUUUACUUAGUCCUAUGAUGUGGAGCACUGUAACUGACCGCUCCACUGAGAA------CAUAUGUUUCCAAUAUUUUCUAAA------CUGAUUAUUGGACAAUAAUAACCA ..........((((.((((((((((((.............)))))))..((((------.((((.......)))).))))...------...))))).))))........... ( -23.92) >DroEre_CAF1 18534 101 + 1 CUUUU-----------UGUCUUGGGGCAUUGAAACUCACCGCUCCCGUGACAAUAGUAACAUUUGUUUCCGUUAUUU-CUAAUUAUUUCUUAGUUAUAAGACAAAAAUAAUCA .((((-----------(((((((((((..((.....))..)))))((.(((((.........)))))..))......-.((((((.....)))))).))))))))))...... ( -19.70) >consensus CUUUUACU_AG_CCUAUGAUGUGGAGCAAUGUAACUCACCGCUCCACUGA_AA____AACAUUUGUUUCCAAUAUUUUCUAAA______CUGAUUAUUGGACAAUAAUAACCA ..........((((.((((((((((((.............)))))))................((....)).....................))))).))))........... (-10.63 = -11.75 + 1.12)


| Location | 7,834,828 – 7,834,926 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 65.05 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -12.50 |
| Energy contribution | -14.83 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
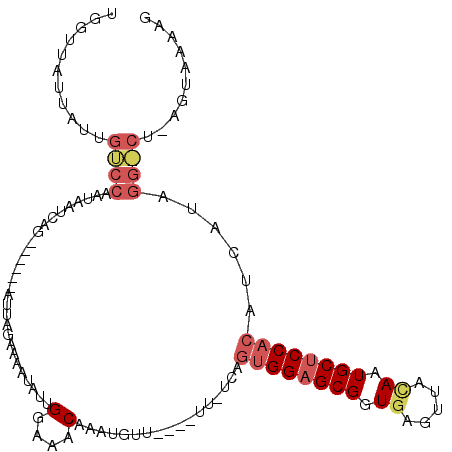
>X_DroMel_CAF1 7834828 98 - 22224390 UGGCUAUUAUUGCCCAAAAAUCAG------CUUAGAAAACAUUGGAAACAAAUGUU---------AGUGGAGCGGUGAGGUAUAUUGCUCCACAUCAUAGGGCUGAGUAAAAG .(((.......)))......((((------(((....(((((((....).))))))---------.(((((((((((.....)))))))))))......)))))))....... ( -34.40) >DroSim_CAF1 16073 101 - 1 UGGUUAUUAUUGUCCAAUAAUCAG------UUUAGAAAAUAUUGGAAACAUAUG------UUCUCAGUGGAGCGGUCAGUUACAGUGCUCCACAUCAUAGGACUAAGUAAAAG ...(((((...((((.........------....((.(((((((....)).)))------)).)).((((((((.(.......).))))))))......))))..)))))... ( -27.00) >DroEre_CAF1 18534 101 - 1 UGAUUAUUUUUGUCUUAUAACUAAGAAAUAAUUAG-AAAUAACGGAAACAAAUGUUACUAUUGUCACGGGAGCGGUGAGUUUCAAUGCCCCAAGACA-----------AAAAG ......((((((((((........((......(((-.((((..(....)...)))).)))...))..(((.(((.((.....)).))))))))))))-----------)))). ( -22.80) >consensus UGGUUAUUAUUGUCCAAUAAUCAG______AUUAGAAAAUAUUGGAAACAAAUGUU____UU_UCAGUGGAGCGGUGAGUUACAAUGCUCCACAUCAUAGG_CU_AGUAAAAG ...........((((............................(....).................((((((((.((.....)).))))))))......)))).......... (-12.50 = -14.83 + 2.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:50 2006