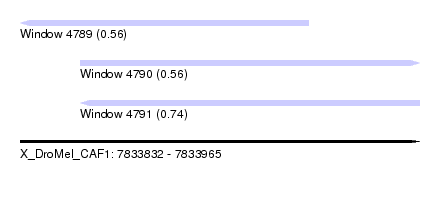
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,833,832 – 7,833,965 |
| Length | 133 |
| Max. P | 0.741316 |

| Location | 7,833,832 – 7,833,928 |
|---|---|
| Length | 96 |
| Sequences | 6 |
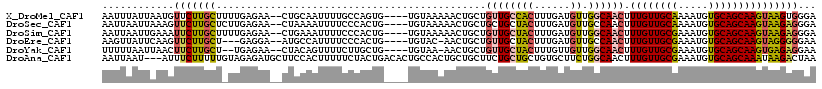
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7833832 96 - 22224390 UGCCAGUG----UGUAAAAACUGCUGUUGCCACUUUGAUGUUGGCAACUUUGUUGCAAAAUGUGCAGCAAGUAAGUGGGA--------------GAAAAUCCGGCGGCGAAAGU .(((....----.........((((((((((((......).)))))))..(((((((.....))))))))))).((.(((--------------.....))).)))))...... ( -32.50) >DroSec_CAF1 9725 96 - 1 UCCCACUG----UGUAAAAACUGCUGCUGCUACUUUGAUGUUGCCAACUUUGUUGCAAAAUGUGCAGCAAGUAAGAGGGA--------------GAAAAUCCGGCGGCGAAAGU ........----..........((....)).(((((..(((((((.((((.((((((.....)))))))))).....(((--------------.....))))))))))))))) ( -25.20) >DroSim_CAF1 15109 96 - 1 UCCCACUG----UGUAAAAACUGCUGUUGCUACUUUGAUGUUGGCAACUUUGUUGCGAAAUGUGCAGCAAGUAAGAGGGA--------------GAAAAUCCGGCGGCGAAAGU ((((.(((----(......))((((((((((((......).)))))))..(((((((.....))))))))))))).))))--------------............((....)) ( -25.40) >DroEre_CAF1 17479 107 - 1 UCCCACUG----UGUAC-AACUGCUGUUGCUACUUUGAUGUUGCCAACUUUGUUGCGAAAUGUGCAGCAAGUAGGGGGAACGAAAAACAAC--AAAAAAUGCGGCGGAGAAAGU ((((.(((----(((((-(((....)))).)))((((.((((.((.((((.((((((.....)))))))))).))(....)....)))).)--)))....)))).)).)).... ( -28.60) >DroYak_CAF1 14038 95 - 1 UCUUGCUG----UGUAA-AACUGCUGUUGCUACUUUGUUGUUGGCAACUUUGUUGCGAAAUGUGCAGCAAGUGAGAGGAA--------------AAAAUGGCGGCGGAGAAAGU ((((((((----(....-.......((((((((......).)))))))(((((((((.....))))))))).........--------------......))))).)))).... ( -25.50) >DroAna_CAF1 6532 110 - 1 UUCUACUGACACUGCCACUGCUGCUUCUGCUGCUGUGCUUCUGGCAACUUUGUUGCGAAAUGUGCAGCAAAUAAGACUAA----AAAAAAUUGCAAAAAUCCGCCGGAGAAAGU .............((.((.((.((....)).)).))))(((((((...(((((((((.....))))))))).........----.....(((.....)))..)))))))..... ( -25.00) >consensus UCCCACUG____UGUAAAAACUGCUGUUGCUACUUUGAUGUUGGCAACUUUGUUGCGAAAUGUGCAGCAAGUAAGAGGAA______________AAAAAUCCGGCGGAGAAAGU ....................(((((((((((((......)).))))))(((((((((.....)))))))))...............................)))))....... (-17.98 = -18.37 + 0.39)


| Location | 7,833,852 – 7,833,965 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7833852 113 + 22224390 UCCCACUUACUUGCUGCACAUUUUGCAACAAAGUUGCCAACAUCAAAGUGGCAACAGCAGUUUUUACA----CACUGGCAAAAUUGCAG--UUCUCAAAAGCAAGAACAUUAAUAAAUU .....(((.(((((((((.(((((((......(((((((.(......))))))))..((((.......----.))))))))))))))))--)......))).))).............. ( -29.60) >DroSec_CAF1 9745 113 + 1 UCCCUCUUACUUGCUGCACAUUUUGCAACAAAGUUGGCAACAUCAAAGUAGCAGCAGCAGUUUUUACA----CAGUGGGAAAAUUUUAG--UUCUCAAGAGCAAGAACUUUAAUUAAUU ((((.((....((((((......(((.((.....((....)).....)).)))))))))((....)).----.)).)))).((((..((--((((........))))))..)))).... ( -27.10) >DroSim_CAF1 15129 113 + 1 UCCCUCUUACUUGCUGCACAUUUCGCAACAAAGUUGCCAACAUCAAAGUAGCAACAGCAGUUUUUACA----CAGUGGGAAAAUUUCAG--UUCUCAAAAGCAAGAAUUUCAAUUAAUU ((((.((.....(((((...(((......)))(((((..((......)).))))).))))).......----.)).)))).((((..((--((((........))))))..)))).... ( -19.72) >DroEre_CAF1 17511 109 + 1 UUCCCCCUACUUGCUGCACAUUUCGCAACAAAGUUGGCAACAUCAAAGUAGCAACAGCAGUU-GUACA----CAGUGGGAAAAUGGCAU--UCCUC---AGCAAGAACUUGAAUAACUU .........((((((((((.....(((((...(.((....)).)......((....)).)))-))...----..)))((((.......)--))).)---)))))).............. ( -23.50) >DroYak_CAF1 14058 110 + 1 UUCCUCUCACUUGCUGCACAUUUCGCAACAAAGUUGCCAACAACAAAGUAGCAACAGCAGUU-UUACA----CAGCAAGAAAACUGUAG--UUCUCA--AGCAAGAAGUUAAUUAAAAA .........((((((((.......))(((...(((((..((......)).))))).((((((-((.(.----......))))))))).)--))....--)))))).............. ( -21.30) >DroAna_CAF1 6562 116 + 1 UUAGUCUUAUUUGCUGCACAUUUCGCAACAAAGUUGCCAGAAGCACAGCAGCAGAAGCAGCAGUGGCAGUGUCAGUAGAAAAAGUGGAAGCAUCUCUACAAAAAGAAAU---AUUAAUU ....((((.((((((((((.(((.(((((...))))).)))..(((.((.((....)).)).)))...))).)))))))....(((((......)))))...))))...---....... ( -28.50) >consensus UCCCUCUUACUUGCUGCACAUUUCGCAACAAAGUUGCCAACAUCAAAGUAGCAACAGCAGUUUUUACA____CAGUGGGAAAAUUGCAG__UUCUCAA_AGCAAGAACUUUAAUAAAUU ....(((((((((((((.......(((((...)))))..........))))))).....((....)).......))))))....................................... (-10.52 = -10.75 + 0.23)

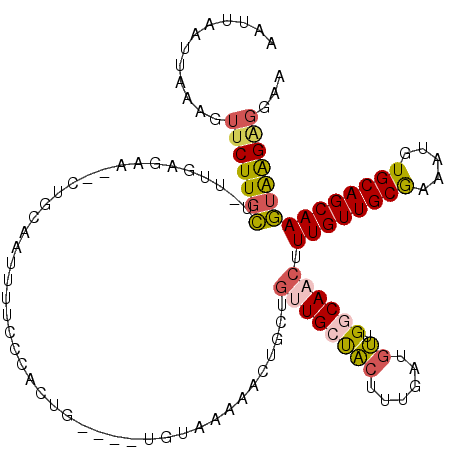
| Location | 7,833,852 – 7,833,965 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7833852 113 - 22224390 AAUUUAUUAAUGUUCUUGCUUUUGAGAA--CUGCAAUUUUGCCAGUG----UGUAAAAACUGCUGUUGCCACUUUGAUGUUGGCAACUUUGUUGCAAAAUGUGCAGCAAGUAAGUGGGA .............(((..(((.......--((((((((((((((((.----.......))))..((((((((......).)))))))......))))))).))))).....)))..))) ( -32.80) >DroSec_CAF1 9745 113 - 1 AAUUAAUUAAAGUUCUUGCUCUUGAGAA--CUAAAAUUUUCCCACUG----UGUAAAAACUGCUGCUGCUACUUUGAUGUUGCCAACUUUGUUGCAAAAUGUGCAGCAAGUAAGAGGGA ..........(((((((......)))))--)).......((((.(((----(......))(((((((((.((.......((((.(((...)))))))...))))))).)))))).)))) ( -27.00) >DroSim_CAF1 15129 113 - 1 AAUUAAUUGAAAUUCUUGCUUUUGAGAA--CUGAAAUUUUCCCACUG----UGUAAAAACUGCUGUUGCUACUUUGAUGUUGGCAACUUUGUUGCGAAAUGUGCAGCAAGUAAGAGGGA ............(((((......)))))--.........((((.(((----(......))((((((((((((......).)))))))..(((((((.....))))))))))))).)))) ( -25.10) >DroEre_CAF1 17511 109 - 1 AAGUUAUUCAAGUUCUUGCU---GAGGA--AUGCCAUUUUCCCACUG----UGUAC-AACUGCUGUUGCUACUUUGAUGUUGCCAACUUUGUUGCGAAAUGUGCAGCAAGUAGGGGGAA ..(.(((((.(((....)))---...))--))).)...(((((....----.((((-(((....)))).)))..........((.((((.((((((.....)))))))))).))))))) ( -28.10) >DroYak_CAF1 14058 110 - 1 UUUUUAAUUAACUUCUUGCU--UGAGAA--CUACAGUUUUCUUGCUG----UGUAA-AACUGCUGUUGCUACUUUGUUGUUGGCAACUUUGUUGCGAAAUGUGCAGCAAGUGAGAGGAA ...........(((((..((--((....--.((((((......))))----))...-..((((..((((.((...((((....))))...)).)))).....))))))))..))))).. ( -34.30) >DroAna_CAF1 6562 116 - 1 AAUUAAU---AUUUCUUUUUGUAGAGAUGCUUCCACUUUUUCUACUGACACUGCCACUGCUGCUUCUGCUGCUGUGCUUCUGGCAACUUUGUUGCGAAAUGUGCAGCAAAUAAGACUAA .......---...((((...(((((((...........)))))))......(((((..((.((.......))...))...)))))..(((((((((.....))))))))).)))).... ( -25.40) >consensus AAUUAAUUAAAGUUCUUGCU_UUGAGAA__CUGCAAUUUUCCCACUG____UGUAAAAACUGCUGUUGCUACUUUGAUGUUGGCAACUUUGUUGCGAAAUGUGCAGCAAGUAAGAGGAA ............(((((((.............................................((((((((......)).)))))).((((((((.....)))))))))))))))... (-17.57 = -17.77 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:47 2006