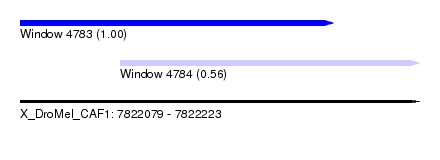
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,822,079 – 7,822,223 |
| Length | 144 |
| Max. P | 0.998256 |

| Location | 7,822,079 – 7,822,192 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -20.17 |
| Energy contribution | -19.07 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
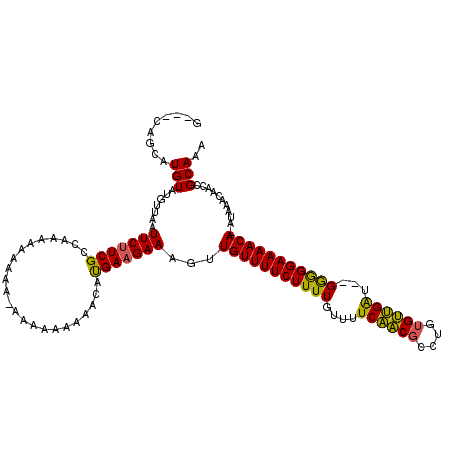
>X_DroMel_CAF1 7822079 113 + 22224390 G---CAGCAUGUAUGUUAAUUCUUCGCCAAAAAAAAA-AAAAAAAAACAUGAAGAAAGUUGUUUUCUUUUGUUUUCAACGCCUGUGUUGAU--GGGGGAAAACA-AUAAACAACCGCAAA .---..((..((.((((..(((((((...........-...........))))))).(((((((((((..(...((((((....)))))))--..)))))))))-)).)))))).))... ( -24.05) >DroSec_CAF1 23912 114 + 1 G---CAUCAUGUAUGUUAAUUCUUCGCCAAAAAAAAAAAAAAAAAAACAUGAAGAAAGUUGUUUUCUUUUGUUUUCAACGCCUGUGUUGAU--GGGGGAAAACA-AUAAACAACCGCAAA (---(..............(((((((.......................))))))).(((((((((((..(...((((((....)))))))--..)))))))))-))........))... ( -23.00) >DroAna_CAF1 24493 107 + 1 GAGAGAGCAUGUAUUCAAAUUCCUCGCCGAAGAA--------ACAGACGCGAUGAAAGUUGUUUUCUUUUGUGUUCGUC-----CGACGGCGGGAAGGAAAACAAAUCAAAACCCGCAAA ....((...(((.(((...((((.(((((..(..--------((..((((((.(((((...)))))..))))))..)).-----)..))))))))).))).)))..))............ ( -29.10) >consensus G___CAGCAUGUAUGUUAAUUCUUCGCCAAAAAAAAA_AAAAAAAAACAUGAAGAAAGUUGUUUUCUUUUGUUUUCAACGCCUGUGUUGAU__GGGGGAAAACA_AUAAACAACCGCAAA .........(((.......(((((((.......................)))))))...(((((((((((....((((((....))))))...)))))))))))...........))).. (-20.17 = -19.07 + -1.10)


| Location | 7,822,115 – 7,822,223 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -17.81 |
| Energy contribution | -16.60 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7822115 108 + 22224390 AAAAAAACAUGAAGAAAGUUGUUUUCUUUUGUUUUCAACGCCUGUGUUGAU--GGGGGAAAACA-AUAAACAACCGCAAAAUGGUUUAUAAAUAACAGGCCCAAGGCAACU .................(((((((((((..(...((((((....)))))))--..)))))))))-))....(((((.....)))))............(((...))).... ( -22.00) >DroSec_CAF1 23949 108 + 1 AAAAAAACAUGAAGAAAGUUGUUUUCUUUUGUUUUCAACGCCUGUGUUGAU--GGGGGAAAACA-AUAAACAACCGCAAAAUGGUUUAUAAAUAACAGGCCCAAGGCAACU .................(((((((((((..(...((((((....)))))))--..)))))))))-))....(((((.....)))))............(((...))).... ( -22.00) >DroAna_CAF1 24527 101 + 1 --ACAGACGCGAUGAAAGUUGUUUUCUUUUGUGUUCGUC-----CGACGGCGGGAAGGAAAACAAAUCAAAACCCGCAAAAUGUUUUAUAAAUAAGAGGCCCAAGGC---C --......(((.(((...((((((((((((.(((.((..-----...))))).)))))))))))).))).....)))....................((((...)))---) ( -25.00) >consensus AAAAAAACAUGAAGAAAGUUGUUUUCUUUUGUUUUCAACGCCUGUGUUGAU__GGGGGAAAACA_AUAAACAACCGCAAAAUGGUUUAUAAAUAACAGGCCCAAGGCAACU ...................(((((((((((....((((((....))))))...)))))))))))..................................(((...))).... (-17.81 = -16.60 + -1.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:40 2006