
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,800,512 – 7,800,669 |
| Length | 157 |
| Max. P | 0.907333 |

| Location | 7,800,512 – 7,800,632 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7800512 120 + 22224390 UAGCAAUCACGUGGAAGUACACCCGGCCCAAAAACCCCACCGGCUUCGAAGCCCAUGGGCAACAUAACCGUGGGCAUAAAGACGAUCUUAUCUAUCGGAGCGUUAUCCGGGAAAAGGGAA .....................(((..(((.............(((((((.((((((((.........))))))))...(((.....))).....)))))))(.....))))....))).. ( -35.70) >DroVir_CAF1 4504 120 + 1 UAGUAUUUGUGGGCGAGGACACCCGGCGGAAAGACGCCGCCGGCAUCGAAGUCGCCCGGCAGCAGCUGCGGCGGCAAAUACGUGAUCUUAUUGAGCGGCGAGUGCCGUGGAAACAGAUUA ..((((((((.(((..((....))((((......))))))).))......((((((((((....)))).)))))))))))).((((((......(((((....)))))(....))))))) ( -52.00) >DroSec_CAF1 3504 120 + 1 UAGCAACGACGUGGAAGGACACCCGGGCCAAACACCCCUCCGGCUUCGAAGCCCAUGGGCAACAUCACCUUGGGCAUAAAGACGAUCUUGUCUAUCGGAGCCUUUUCCGGGAACAGGUUA ((((.....(.((((((((.....(((........))))))((((((((.(((((.((.........)).)))))....(((((....))))).))))))))..))))).).....)))) ( -41.60) >DroSim_CAF1 4018 120 + 1 UAGCAACGACGUGGAAGGACACCCGGGCCAAACACCCCGCCGGCUUCGAAGCCCAUGGGCAACAUCACCUUGGGCAUAAAGACGAUCUUGUCUAUCGGAGUCUUUUCCGGGAACAGUUUA .........(.(((((((.....((((........))))..((((((((.(((((.((.........)).)))))....(((((....))))).))))))))))))))).)......... ( -39.00) >DroEre_CAF1 3475 120 + 1 UAGGAACUCGGUGGCAGAACACCCGGGCCAAAAACGCCUCCGGCUUCAAAGCCCAUGGGCAACAUCACCGUGGGCAUGAAGACGAUCUUGUCCAGCGGCACCUCACCCGGGAACAGGGAA .(((...(((.(((((((......((((.......)))).((.(((((..((((((((.........)))))))).))))).)).)).)).))).)))..)))..((((....).))).. ( -41.60) >DroYak_CAF1 4010 120 + 1 UAGGAACGCCGUGGAAGAACACCAGGGCCAAAUACCCCGCCGGCUUCGAAGCCCAUGGGCAACAUGACCGUGGGCAUGAAGACGAACUUGUCCAUCGGUACCUCACCCGGGAACAGGGAA .(((...((((((((.........(((........)))..((.(((((..((((((((.........)))))))).))))).))......)))).)))).)))..((((....).))).. ( -42.00) >consensus UAGCAACGACGUGGAAGGACACCCGGGCCAAAAACCCCGCCGGCUUCGAAGCCCAUGGGCAACAUCACCGUGGGCAUAAAGACGAUCUUGUCUAUCGGAGCCUUACCCGGGAACAGGGAA .....................((((((........))).(((........((((((((.........)))))))).....((((....))))...)))..........)))......... (-23.64 = -24.00 + 0.36)



| Location | 7,800,552 – 7,800,669 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -45.93 |
| Consensus MFE | -29.56 |
| Energy contribution | -30.95 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7800552 117 + 22224390 CGGCUUCGAAGCCCAUGGGCAACAUAACCGUGGGCAUAAAGACGAUCUUAUCUAUCGGAGCGUUAUCCGGGAAAAGGGAAGCCGGCAAAUCGAAGCAGGGCGCCUGGAAGCCCUCAA (((((((...((((((((.........))))))))..........((((.(((..((((......))))))).)))))))))))((........))(((((........)))))... ( -47.00) >DroSec_CAF1 3544 117 + 1 CGGCUUCGAAGCCCAUGGGCAACAUCACCUUGGGCAUAAAGACGAUCUUGUCUAUCGGAGCCUUUUCCGGGAACAGGUUAGUCGGCAAAUCGAAGCAGGGCGUCUGGAAGCCCUCAA ..(((((((.(((((.((.........)).))))).....(((...(((((...((((((....))))))..)))))...)))......)))))))(((((........)))))... ( -47.60) >DroSim_CAF1 4058 117 + 1 CGGCUUCGAAGCCCAUGGGCAACAUCACCUUGGGCAUAAAGACGAUCUUGUCUAUCGGAGUCUUUUCCGGGAACAGUUUAGUCGGCAAAUCGAAGCAGGGCGUCUGGAAGCCCUCGA (((((((((.(((((.((.........)).))))).....(((....((((...((((((....))))))..))))....)))......)))))))(((((........))))))). ( -45.00) >DroEre_CAF1 3515 117 + 1 CGGCUUCAAAGCCCAUGGGCAACAUCACCGUGGGCAUGAAGACGAUCUUGUCCAGCGGCACCUCACCCGGGAACAGGGAAGCCGGCACCUCGAAGCAGGGCGUCUGGAAGCCCUCGA .((((((...((((((((.........))))))))....(((((.((((((...(.((..((...((((....).))).....))..)).)...))))))))))).))))))..... ( -47.70) >DroYak_CAF1 4050 117 + 1 CGGCUUCGAAGCCCAUGGGCAACAUGACCGUGGGCAUGAAGACGAACUUGUCCAUCGGUACCUCACCCGGGAACAGGGAAGCGGGCAUCUCGAAGCAGGGCGUCUGGAAGCCCUCGA (((((((((.((((((((.........))))))))(((....((..(((((((...(((.....)))..)).)))))....))..))).)))))))(((((........))))))). ( -47.90) >DroAna_CAF1 3708 117 + 1 CAGCAUCGAAUCCGGACGGCAGGAGCUGGCGCGGCAAAUAGGUGAUCUUGCCCAGGGGGGCGUGCAUGGGGAACAUGGUGCGCGGCAAGUCGAAGCAGAUGCACUUCUCGUCCUGGA ..........(((((((((.(((.((..((.((((..............((((....))))((((((.(....)...)))))).....))))..))....)).))).)))))).))) ( -40.40) >consensus CGGCUUCGAAGCCCAUGGGCAACAUCACCGUGGGCAUAAAGACGAUCUUGUCCAUCGGAGCCUUAUCCGGGAACAGGGAAGCCGGCAAAUCGAAGCAGGGCGUCUGGAAGCCCUCGA ..(((((((.(((((.((.........)).)))))...........(((((((..(((........))))).)))))............)))))))(((((........)))))... (-29.56 = -30.95 + 1.39)

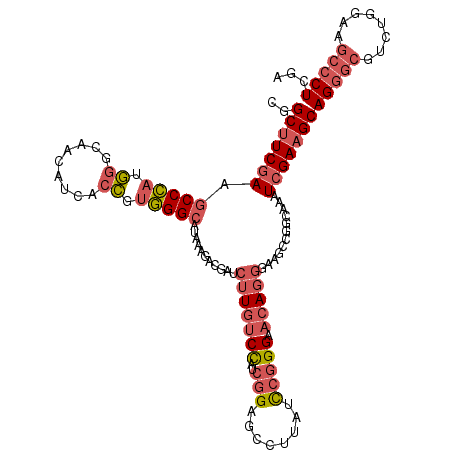
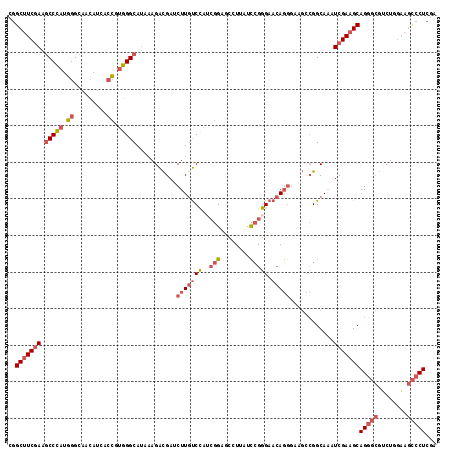
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:33 2006