
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,796,079 – 7,796,192 |
| Length | 113 |
| Max. P | 0.705425 |

| Location | 7,796,079 – 7,796,192 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7796079 113 - 22224390 UCGCACUCUAAAUCGGUGAGAGUAACGCACAGCUGAACCACAAUUUGAUAGUACAGCAGAUUGUACAUGACUUUCACCAUGCACGUAAAGUAAUCUAAAUGG-GAUAAAAAAAA----- ((.((.........(((((((((...((...))......((((((((.........)))))))).....))))))))).(((.......))).......)).-)).........----- ( -20.80) >DroVir_CAF1 16308 115 - 1 UCGCACUCGAAAUCGGUGAGCGUUACGCACAAUUGCACAACAAUCUGAUAGUAGAGCAGAUUGUACAUGACCUUUACCAUGCAGGUGAAAUAAUCUGUAACG-AACA---CGUAGAUGA ......(((.....((((((.((((.(((....)))...((((((((.........))))))))...)))).)))))).(((((((......))))))).))-)...---......... ( -29.30) >DroGri_CAF1 19013 111 - 1 UCGCACUCGAGUUCGGUGAGCGUGACGCACAAUUGGACAACAAUCUGAUAGUAGAGCAGAUUGUACAUCACCUUAACCAUGCAGGUGAAAUAAUCUGCAAUACAACA---AAUA----- ((((.((((.......)))).))))......((..((......))..)).(((..(((((((((...((((((.........)))))).)))))))))..)))....---....----- ( -29.90) >DroWil_CAF1 12777 112 - 1 UCACACUCGAGAUCAGUCAGGGUGACGCAAAGUUGGACUACAAUCUGAUAGUAUAACAGAUUAUACAUGACUUUAACAAUGCAGGUGAAGUAGUCUGCAAGA-AUCA-UAGACA----- ...............(((...((((..((((((((...((.((((((.........)))))).))..))))))).....((((((........)))))).).-.)))-).))).----- ( -21.40) >DroYak_CAF1 16741 111 - 1 UCGCACUCAAAAUCCGUGAGGGUAACGCACAGCUGAACCACAAUCUGGUAGUACAGCAGGUUGUACAUGACUUUCACCAUGCAAGUGAAGUAAUCUAAAUGG-CAAG--CGAAA----- ((((.......((((....))))...((.((...((((((.....)))).((((((....))))))...((((.(((.......)))))))..))....)))-)..)--)))..----- ( -28.20) >DroAna_CAF1 15267 107 - 1 UCGACCUCGAGAUCCGUGAGGGUGACGCAAAGCUGGACUACGAUCUGGUAGUAUAGCAGGUUGUACAUCACCUUCACCAUGCAGGUGAAAUAAUCUGCG-----AAG--GAAAA----- (((....)))..(((((((((((((.((((.((((.(((((......))))).))))...))))...))))))))))..(((((((......)))))))-----..)--))...----- ( -42.10) >consensus UCGCACUCGAAAUCGGUGAGGGUGACGCACAGCUGGACAACAAUCUGAUAGUACAGCAGAUUGUACAUGACCUUCACCAUGCAGGUGAAAUAAUCUGCAAGG_AACA___AAAA_____ ...............((((((((....((....))....((((((((.........)))))))).....))))))))..(((((((......))))))).................... (-15.23 = -15.95 + 0.72)

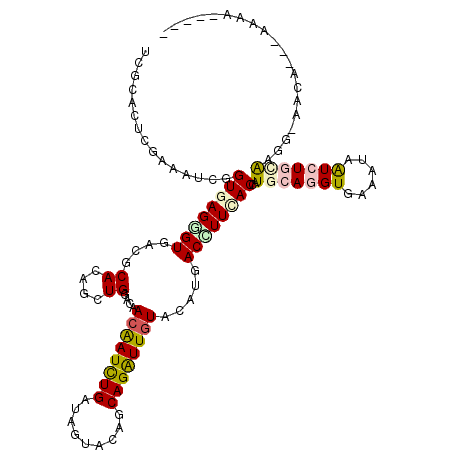
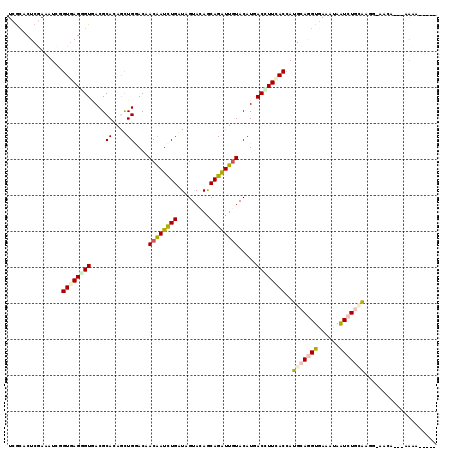
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:26 2006