| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 966,328 – 966,420 |
| Length | 92 |
| Max. P | 0.843096 |

| Location | 966,328 – 966,420 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.08 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -22.93 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 966328 92 + 22224390 --GAAG---------GGGAAAAGCGCAGGAAAAACAGA-AGGCAGCUC-GCUCGCCUUCUGUUAGCUGUGGAUGGCUGUUAGGGAAAAAGUCUGCCAAGACGAGG --..((---------.((.....(((((....((((((-((((((...-.)).))))))))))..))))).....)).)).........((((....)))).... ( -27.60) >DroSec_CAF1 49236 92 + 1 --GAAG---------GGGAAAAGCGCAGGAAAAACAGA-AGGCAGCUC-GCUCGCCUUCUGUUAGCUGUGGAUGGCUGUUAGGGAAAAAGUCUGCCAAGACGAGG --..((---------.((.....(((((....((((((-((((((...-.)).))))))))))..))))).....)).)).........((((....)))).... ( -27.60) >DroYak_CAF1 53746 105 + 1 GAAAAGGGGAAAAGUGGGAAAAGCGCAGGAAAAACAGAAAGGCAGGGCAGCUCGCCUUCUGUUUGCUGUGAAUGGCUGUUAGGGAAAAAGUCUGCCAAGACGAGG ............(((.(......(((((...((((((((.(((..(....)..))))))))))).)))))..).)))............((((....)))).... ( -26.30) >consensus __GAAG_________GGGAAAAGCGCAGGAAAAACAGA_AGGCAGCUC_GCUCGCCUUCUGUUAGCUGUGGAUGGCUGUUAGGGAAAAAGUCUGCCAAGACGAGG ................((.....(((((....((((((.((((((.....)).))))))))))..))))).....))............((((....)))).... (-22.93 = -22.93 + 0.00)


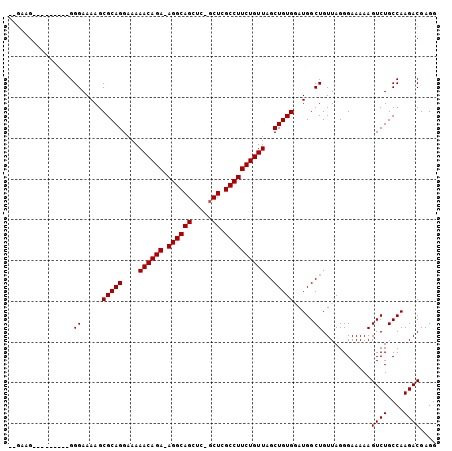
| Location | 966,328 – 966,420 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.08 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -18.53 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 966328 92 - 22224390 CCUCGUCUUGGCAGACUUUUUCCCUAACAGCCAUCCACAGCUAACAGAAGGCGAGC-GAGCUGCCU-UCUGUUUUUCCUGCGCUUUUCCC---------CUUC-- ..........((((..............(((........)))((((((((((.((.-...))))))-))))))....)))).........---------....-- ( -23.90) >DroSec_CAF1 49236 92 - 1 CCUCGUCUUGGCAGACUUUUUCCCUAACAGCCAUCCACAGCUAACAGAAGGCGAGC-GAGCUGCCU-UCUGUUUUUCCUGCGCUUUUCCC---------CUUC-- ..........((((..............(((........)))((((((((((.((.-...))))))-))))))....)))).........---------....-- ( -23.90) >DroYak_CAF1 53746 105 - 1 CCUCGUCUUGGCAGACUUUUUCCCUAACAGCCAUUCACAGCAAACAGAAGGCGAGCUGCCCUGCCUUUCUGUUUUUCCUGCGCUUUUCCCACUUUUCCCCUUUUC ..........((((...............((........))((((((((((((........)))).))))))))...))))........................ ( -19.50) >consensus CCUCGUCUUGGCAGACUUUUUCCCUAACAGCCAUCCACAGCUAACAGAAGGCGAGC_GAGCUGCCU_UCUGUUUUUCCUGCGCUUUUCCC_________CUUC__ ..........((((...............((........)).((((((((((.((.....)))))).))))))....))))........................ (-18.53 = -18.53 + 0.00)

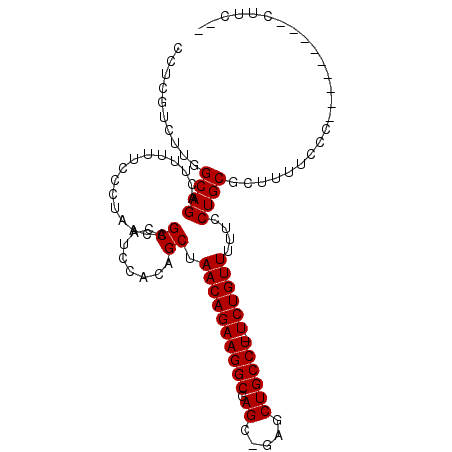
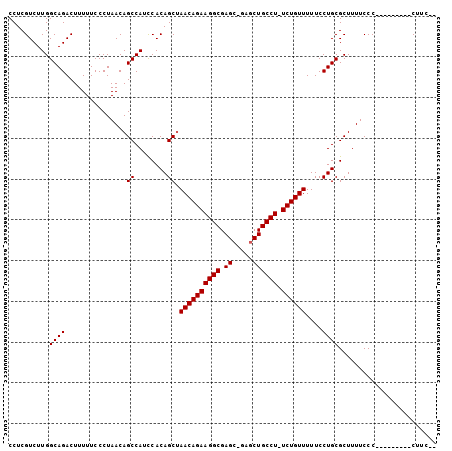
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:56 2006