
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,772,650 – 7,772,780 |
| Length | 130 |
| Max. P | 0.990342 |

| Location | 7,772,650 – 7,772,755 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -48.63 |
| Consensus MFE | -22.38 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
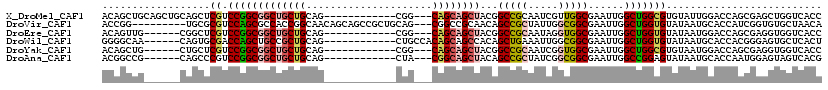
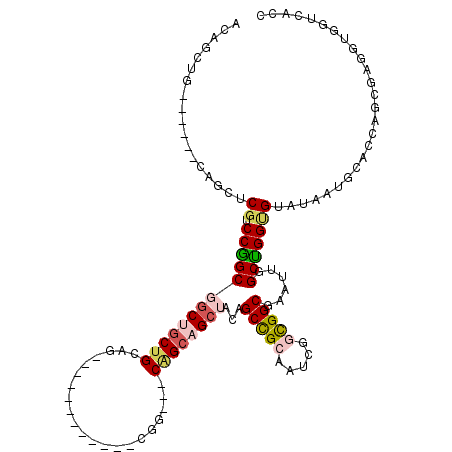
>X_DroMel_CAF1 7772650 105 + 22224390 ACAGCUGCAGCUGCAGCUCGUCCGGCGGCUGCUGCAG------------CGG---CAGCAGCUACGGCCGCAAUCGUUGGCGAAUUGGCUGGCGUGUAUUGGACCAGCGAGCUGGUCACC ..((((((....))))))((.(((..(((((((((..------------..)---)))))))).))).))(((((((..((......))..)))...))))((((((....))))))... ( -53.00) >DroVir_CAF1 197 108 + 1 ACCGG---------UGCGCGUCCAGCGCCACCGGCAACAGCAGCCGCUGCAG---CGGCCGCAACAGCCGCUAUUGGCGGCGAAUUGGCUGGUGUAUAAUGCACCAUCGGUGUGCUAACA .((((---------(((((.....))).))))))....((((((((((..((---((((.......))))))...))))))..(((((.((((((.....))))))))))).)))).... ( -52.70) >DroEre_CAF1 5280 99 + 1 ACAGUUG------CGGCUCGUCCGGCGGCUGCUGCAG------------CGG---CAGCAGCUACGGCCGCAAUAGGUGGCGAAUUGGCUGGUGUAUAAUGGACCAGCGAGGUGGUCACC ...((((------((((.(((.....(((((((((..------------..)---))))))))))))))))))).((((((.(.((.((((((.(.....).)))))).)).).)))))) ( -54.10) >DroWil_CAF1 203 102 + 1 GGGGCAA------CAGUGCGACCAGCUGCCGCUGCAG------------CUGCCACAGCAGCCACAGCUGAAAUUGGCGGCGAAUUGGCUGGUGUAUAAUGCACCACGGGAGUGCUCACU .((((..------..(((((..(((((((....))))------------)))..(((.((((((..((((.......))))....)))))).)))....))))).((....))))))... ( -42.30) >DroYak_CAF1 203 99 + 1 ACAGCUG------CUGCUCGUCCGGCGGCUGCUGCAG------------CGG---CAGCAGCUACGGCCGCAAUCGGUGGCGAAUUGGCUGGCGUGUAAUGGACCAGCGAGGUGGUCACC ..(((((------((((.(((.((((....))))..)------------)))---))))))))..((((((..(((....))).((.((((((........).))))).))))))))... ( -45.20) >DroAna_CAF1 6063 99 + 1 ACGGCCG------CAGCCCGUCCGGCGGCUGCUGCAG------------CUA---CGGCAGCUACAGCCGCUAUCGGCGGCGAAUUGGCCGGAGUAUAAUGCACCAAUGGAGUAGUCACG .((((((------...((((((.((((((((..((.(------------(..---..)).))..))))))))...))))).)...))))))..((....(((.((...)).)))...)). ( -44.50) >consensus ACAGCUG______CAGCUCGUCCGGCGGCUGCUGCAG____________CGG___CAGCAGCUACAGCCGCAAUCGGCGGCGAAUUGGCUGGUGUAUAAUGCACCAGCGAGGUGGUCACC ..................((.(((((((((((((.....................))))))))...(((((.....)))))......))))))).......................... (-22.38 = -23.02 + 0.64)
| Location | 7,772,650 – 7,772,755 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -44.33 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7772650 105 - 22224390 GGUGACCAGCUCGCUGGUCCAAUACACGCCAGCCAAUUCGCCAACGAUUGCGGCCGUAGCUGCUG---CCG------------CUGCAGCAGCCGCCGGACGAGCUGCAGCUGCAGCUGU (....)((((((((((((....(((..(((.((....(((....)))..))))).)))(((((((---(..------------..)))))))).))))).)))))))((((....)))). ( -47.20) >DroVir_CAF1 197 108 - 1 UGUUAGCACACCGAUGGUGCAUUAUACACCAGCCAAUUCGCCGCCAAUAGCGGCUGUUGCGGCCG---CUGCAGCGGCUGCUGUUGCCGGUGGCGCUGGACGCGCA---------CCGGU .........((((.(((((.......))))).......(((((.((((((((((((((((((...---)))))))))))))))))).)))))((((.....)))).---------.)))) ( -54.60) >DroEre_CAF1 5280 99 - 1 GGUGACCACCUCGCUGGUCCAUUAUACACCAGCCAAUUCGCCACCUAUUGCGGCCGUAGCUGCUG---CCG------------CUGCAGCAGCCGCCGGACGAGCCG------CAACUGU (((((.......((((((.........))))))....))))).....((((((((((.(((((((---(..------------..))))))))......))).))))------))).... ( -42.70) >DroWil_CAF1 203 102 - 1 AGUGAGCACUCCCGUGGUGCAUUAUACACCAGCCAAUUCGCCGCCAAUUUCAGCUGUGGCUGCUGUGGCAG------------CUGCAGCGGCAGCUGGUCGCACUG------UUGCCCC ((((.(((((.....))))).......((((((......(((((......(((((((.((....)).))))------------)))..))))).))))))..)))).------....... ( -39.50) >DroYak_CAF1 203 99 - 1 GGUGACCACCUCGCUGGUCCAUUACACGCCAGCCAAUUCGCCACCGAUUGCGGCCGUAGCUGCUG---CCG------------CUGCAGCAGCCGCCGGACGAGCAG------CAGCUGU (((....)))..(((.((((.......(((.((....(((....)))..))))).(..(((((((---(..------------..))))))))..).)))).)))..------....... ( -39.60) >DroAna_CAF1 6063 99 - 1 CGUGACUACUCCAUUGGUGCAUUAUACUCCGGCCAAUUCGCCGCCGAUAGCGGCUGUAGCUGCCG---UAG------------CUGCAGCAGCCGCCGGACGGGCUG------CGGCCGU .(((((((......)))).))).......(((((.....(((((.....)))))((((((((...---)))------------)))))((((((........)))))------)))))). ( -42.40) >consensus GGUGACCACCCCGCUGGUCCAUUAUACACCAGCCAAUUCGCCACCGAUUGCGGCCGUAGCUGCUG___CCG____________CUGCAGCAGCCGCCGGACGAGCUG______CAGCUGU .((((.......((((((.........))))))....)))).........((((.((.(((((......................))))).)).))))...................... (-20.86 = -21.17 + 0.31)


| Location | 7,772,687 – 7,772,780 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.13 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7772687 93 + 22224390 ---------CGG---CAGCAGCUACGGCCGCAAUCGUUGGCGAAUUGGCUGGCGUGUAUUGGACCAGCGAGCUGGUCACCGCAGCCGA------CGGAACAUACUUGUUG---C ---------(((---(.((.((....)).))...(((..((......))..)))(((....((((((....))))))...))))))).------.(.((((....)))).---) ( -36.50) >DroVir_CAF1 228 102 + 1 CAGCCGCUGCAG---CGGCCGCAACAGCCGCUAUUGGCGGCGAAUUGGCUGGUGUAUAAUGCACCAUCGGUGUGCUAACACCAGCCGG---------UGCAUAUUCCUUAAAGG ..((((((..((---((((.......))))))...))))))(.((((((((((((.((..(((((...)))))..)))))))))))))---------).).............. ( -49.10) >DroEre_CAF1 5311 93 + 1 ---------CGG---CAGCAGCUACGGCCGCAAUAGGUGGCGAAUUGGCUGGUGUAUAAUGGACCAGCGAGGUGGUCACCGCAGCCGG------UGCUGCAUACUUGUUG---C ---------(((---(((((((..((((.((....((((((.(.((.((((((.(.....).)))))).)).).)))))))).)))).------.))))).....)))))---. ( -42.30) >DroWil_CAF1 234 98 + 1 ---------CUGCCACAGCAGCCACAGCUGAAAUUGGCGGCGAAUUGGCUGGUGUAUAAUGCACCACGGGAGUGCUCACUGUUGCCGGUUGCUGUGGGGAAUAUUUG------- ---------...(((((((((((...(((......)))(((((..((((.(((((.....)))))((....)))).))...))))))))))))))))..........------- ( -43.20) >DroYak_CAF1 234 99 + 1 ---------CGG---CAGCAGCUACGGCCGCAAUCGGUGGCGAAUUGGCUGGCGUGUAAUGGACCAGCGAGGUGGUCACCGCAGCCGGUGGCGGUGGUGCAUACUUGUUG---C ---------..(---(((((((.((.(((((.((((((.((......((((((........).)))))..(((....))))).)))))).))))).)))).....)))))---) ( -43.60) >DroMoj_CAF1 7188 102 + 1 CGGCCGCCGCAG---CAGCGGCCACAGCGGCUAUUGGUGGUGAAUUGGCCGGCGUAUAGUGCACCAUCGGUGUGGUGACACCAGCGGG---------UGCAUACUCCUUGAAUG .(((((((((..---..)))))..((.((.(....).)).))....))))((.((...(((((((....(.(((....))))....))---------))))))).))....... ( -42.90) >consensus _________CGG___CAGCAGCUACAGCCGCAAUUGGUGGCGAAUUGGCUGGCGUAUAAUGCACCAGCGAGGUGGUCACCGCAGCCGG______UGGUGCAUACUUGUUG___C ...................(((((..(((((.....)))))....)))))(((........((((((....))))))......)))............................ (-18.02 = -18.52 + 0.50)


| Location | 7,772,687 – 7,772,780 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7772687 93 - 22224390 G---CAACAAGUAUGUUCCG------UCGGCUGCGGUGACCAGCUCGCUGGUCCAAUACACGCCAGCCAAUUCGCCAACGAUUGCGGCCGUAGCUGCUG---CCG--------- (---((.((..((((..(((------(.(((((((((((((((....))))))....)).)).)))))...(((....)))..)))).))))..)).))---)..--------- ( -32.70) >DroVir_CAF1 228 102 - 1 CCUUUAAGGAAUAUGCA---------CCGGCUGGUGUUAGCACACCGAUGGUGCAUUAUACACCAGCCAAUUCGCCGCCAAUAGCGGCUGUUGCGGCCG---CUGCAGCGGCUG .......((........---------))(((((((((..((((.......)))).....))))))))).....(((((...(((((((((...))))))---)))..))))).. ( -47.30) >DroEre_CAF1 5311 93 - 1 G---CAACAAGUAUGCAGCA------CCGGCUGCGGUGACCACCUCGCUGGUCCAUUAUACACCAGCCAAUUCGCCACCUAUUGCGGCCGUAGCUGCUG---CCG--------- .---......(((.(((((.------.(((((((((((........((((((.........))))))........))))....)))))))..)))))))---)..--------- ( -36.89) >DroWil_CAF1 234 98 - 1 -------CAAAUAUUCCCCACAGCAACCGGCAACAGUGAGCACUCCCGUGGUGCAUUAUACACCAGCCAAUUCGCCGCCAAUUUCAGCUGUGGCUGCUGUGGCAG--------- -------..........((((((((...(((...(((....)))....(((((.......)))))))).....(((((...........)))))))))))))...--------- ( -31.30) >DroYak_CAF1 234 99 - 1 G---CAACAAGUAUGCACCACCGCCACCGGCUGCGGUGACCACCUCGCUGGUCCAUUACACGCCAGCCAAUUCGCCACCGAUUGCGGCCGUAGCUGCUG---CCG--------- (---((.......))).....((.((.((((((((((.........((((((.........)))))).....(((........))))))))))))).))---.))--------- ( -32.20) >DroMoj_CAF1 7188 102 - 1 CAUUCAAGGAGUAUGCA---------CCCGCUGGUGUCACCACACCGAUGGUGCACUAUACGCCGGCCAAUUCACCACCAAUAGCCGCUGUGGCCGCUG---CUGCGGCGGCCG .......((.(((((((---------((...((((((....))))))..))))))...))).))((........)).......(((((((..((....)---)..))))))).. ( -41.20) >consensus G___CAACAAGUAUGCACCA______CCGGCUGCGGUGACCACCCCGCUGGUCCAUUAUACACCAGCCAAUUCGCCACCAAUUGCGGCCGUAGCUGCUG___CCG_________ ............................(((((..(((((((......)))))......))..)))))...............(((((....)))))................. (-13.74 = -14.68 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:21 2006