
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,772,058 – 7,772,169 |
| Length | 111 |
| Max. P | 0.977786 |

| Location | 7,772,058 – 7,772,169 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -11.69 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
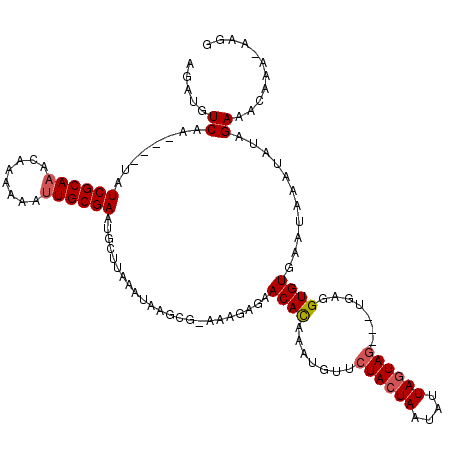
>X_DroMel_CAF1 7772058 111 + 22224390 AGUUGUCAA----UAUCGCAAACAAAAAAUUGCGAAUGCUUAAAUAAGCG-AAAGAGAACACAAAUGUUCUACUAAUAUUAGUAG---UGAGGUGUGAAUAAAUAUAGAAACAAA-AAGG .(((.((..----..((((((........)))))).(((((....)))))-...)).)))(((......((((((....))))))---.....)))...................-.... ( -21.40) >DroSec_CAF1 6174 104 + 1 GGGUGUCAA----UAUCGCAAACAAAAAAUUGCGAA--------UAAGCGGAAAGAGAACAUAAAUGUUCUACUAAUAUUAGUAG---UGAGGUGUGAAUAAAUAUAGAAACAAA-AAGG ..((.((..----..((((((........)))))).--------...............((((......((((((....))))))---.....))))..........)).))...-.... ( -16.20) >DroYak_CAF1 6374 118 + 1 AGAUGUCAAAUUGUAUCGCAAACACA-AACUGCGAAUGCUUAAAUAAGCA-AAAGAGAACACAAAAGUACUAAUAAUAGUAGUAAGCUGGGGGUGUUGAGAAAUAUAGAAACAAAAAAGG ...(((.....(((.((...(((((.-..(.((...(((((....)))))-..............(.(((((....))))).)..)).)...)))))..)).))).....)))....... ( -18.40) >consensus AGAUGUCAA____UAUCGCAAACAAAAAAUUGCGAAUGCUUAAAUAAGCG_AAAGAGAACACAAAUGUUCUACUAAUAUUAGUAG___UGAGGUGUGAAUAAAUAUAGAAACAAA_AAGG .....((........((((((........)))))).......................((((.......((((((....)))))).......))))...........))........... (-11.69 = -12.47 + 0.78)

| Location | 7,772,058 – 7,772,169 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -11.81 |
| Energy contribution | -12.04 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
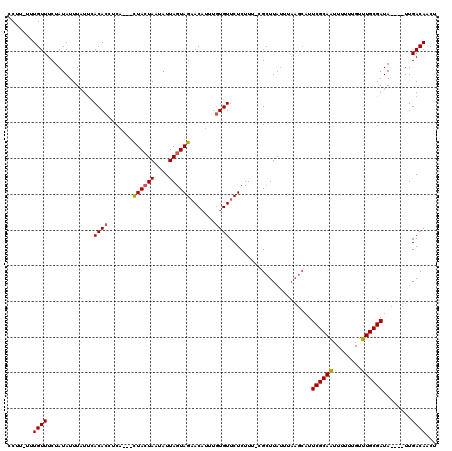
>X_DroMel_CAF1 7772058 111 - 22224390 CCUU-UUUGUUUCUAUAUUUAUUCACACCUCA---CUACUAAUAUUAGUAGAACAUUUGUGUUCUCUUU-CGCUUAUUUAAGCAUUCGCAAUUUUUUGUUUGCGAUA----UUGACAACU ....-.(((((..((........((((.....---((((((....))))))......))))........-.((((....))))..((((((........))))))))----..))))).. ( -19.10) >DroSec_CAF1 6174 104 - 1 CCUU-UUUGUUUCUAUAUUUAUUCACACCUCA---CUACUAAUAUUAGUAGAACAUUUAUGUUCUCUUUCCGCUUA--------UUCGCAAUUUUUUGUUUGCGAUA----UUGACACCC ....-..((((..((.................---.(((((....)))))(((((....)))))............--------.((((((........))))))))----..))))... ( -13.60) >DroYak_CAF1 6374 118 - 1 CCUUUUUUGUUUCUAUAUUUCUCAACACCCCCAGCUUACUACUAUUAUUAGUACUUUUGUGUUCUCUUU-UGCUUAUUUAAGCAUUCGCAGUU-UGUGUUUGCGAUACAAUUUGACAUCU .....................((((..............(((((....)))))...(((((........-(((((....))))).((((((..-.....))))))))))).))))..... ( -16.00) >consensus CCUU_UUUGUUUCUAUAUUUAUUCACACCUCA___CUACUAAUAUUAGUAGAACAUUUGUGUUCUCUUU_CGCUUAUUUAAGCAUUCGCAAUUUUUUGUUUGCGAUA____UUGACAACU .......((((.............((((.......((((((....)))))).......)))).......................((((((........))))))........))))... (-11.81 = -12.04 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:17 2006