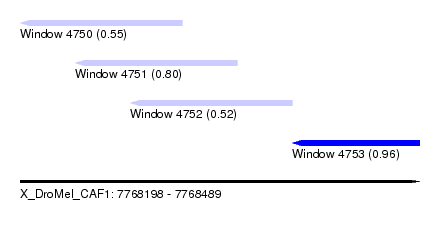
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,768,198 – 7,768,489 |
| Length | 291 |
| Max. P | 0.958138 |

| Location | 7,768,198 – 7,768,316 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 98.87 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7768198 118 - 22224390 CAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCCAUUGUGAUAACAGCUGCGUAAACGAAACACGUUAGAACCU .(((((.((((((((((...(((.....)))...))))))))...........((((((....(((....)))....)))))).......))))))).((((.....))))....... ( -20.30) >DroSec_CAF1 2309 118 - 1 CAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCCAUUGUGAUAACAUCUGCAUAAACGAAACACGUUAGAACCU .(((((...((((((((...(((.....)))...))))))))...........((((((....(((....)))....)))))).........))))).((((.....))))....... ( -18.00) >DroSim_CAF1 2331 118 - 1 CAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCCAUUGUGAUAACAGCUGCGUAAACGAAACACGUUAGAACCU .(((((.((((((((((...(((.....)))...))))))))...........((((((....(((....)))....)))))).......))))))).((((.....))))....... ( -20.30) >consensus CAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCCAUUGUGAUAACAGCUGCGUAAACGAAACACGUUAGAACCU .(((((.((((((((((...(((.....)))...))))))))...........((((((....(((....)))....)))))).......))))))).((((.....))))....... (-18.55 = -18.67 + 0.11)

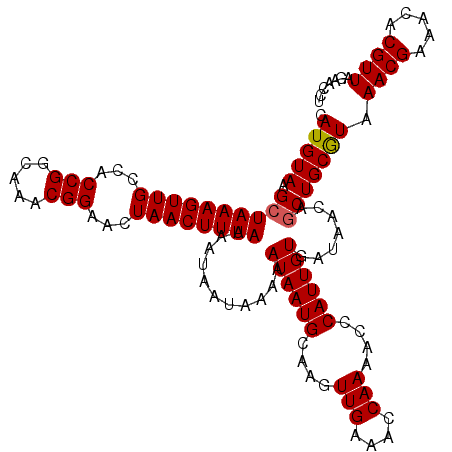
| Location | 7,768,238 – 7,768,356 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -19.71 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7768238 118 - 22224390 UAAACACUAGAAUCGUGGGGCAAAUAACAUUUAGCUGCUGCAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCC ...............((((((((......(((((((((.....)).))))))).))))).(((.....)))......................................)))...... ( -21.50) >DroSec_CAF1 2349 111 - 1 UAAACACCAGAAUCGUGGGGCAAAUAA-------CUGCUCCAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCC ..(((........(((((((((.....-------.))))))))).....((((((((...(((.....)))...))))))))....................)))............. ( -23.50) >DroSim_CAF1 2371 118 - 1 UAAACACCAGAAUUGUGGGGCAAAUAACAUUUAGCUGCUCCAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCC ............(((((((((((......(((((((((.....)).))))))).))))).)).))))..((...((((((....................))))))...))....... ( -21.65) >consensus UAAACACCAGAAUCGUGGGGCAAAUAACAUUUAGCUGCUCCAUGUAAGCUAAAGUUGCCACCGGCAAACGGAACUAACUUUAAAAUAAUAAAAAUAAUGCAAGUUGAAACCAAAACCC ..(((........(((((((((.............))))))))).....((((((((...(((.....)))...))))))))....................)))............. (-19.71 = -19.82 + 0.11)

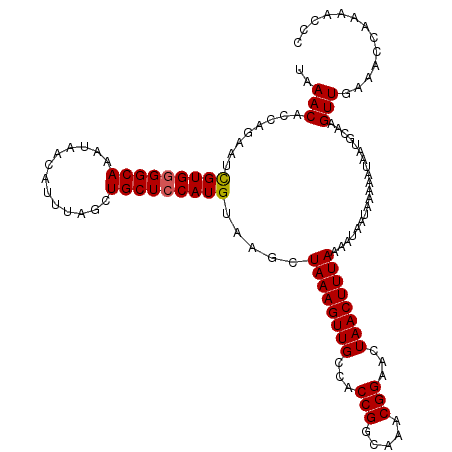

| Location | 7,768,278 – 7,768,396 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7768278 118 - 22224390 GCGGCUUUUGAGGGUUCUGAAAACAAAAUAUGGAAACCAUUAAACACUAGAAUCGUGGGGCAAAUAACAUUUAGCUGCUGCAUGUAAGCUAAAG--UUGCCACCGGCAAACGGAACUAAC ............(((((((............(....).................(((((((((......(((((((((.....)).))))))).--))))).)).))...)))))))... ( -26.80) >DroSec_CAF1 2389 111 - 1 GCGGCUUUUGAGGGUUCUGAAAACAAAAUAUGGAAACUAUUAAACACCAGAAUCGUGGGGCAAAUAA-------CUGCUCCAUGUAAGCUAAAG--UUGCCACCGGCAAACGGAACUAAC ((((((((.(..(((((((.......((((.(....)))))......)))))))((((((((.....-------.)))))))).....).))))--))))..(((.....)))....... ( -33.22) >DroSim_CAF1 2411 118 - 1 GCGGCUUUUGAGGGUUCUGAAAACAAAAUAUGGAAACCAUUAAACACCAGAAUUGUGGGGCAAAUAACAUUUAGCUGCUCCAUGUAAGCUAAAG--UUGCCACCGGCAAACGGAACUAAC ((((((((.(..(((((((..........((((...)))).......)))))))((((((((..((.....))..)))))))).....).))))--))))..(((.....)))....... ( -29.33) >DroYak_CAF1 2482 120 - 1 GCGUCUUUUGAGGGUUCUAAAAACAAAAUAAGAAAACCAUUAAACACCAGACUUGUUGGGCAAUUAACAUUUAACUGCUGGAUGUAGGCUAAAUUUUUGCCAUCGGCAGACGGAACUAAC .(((((.((((((.((((............))))..)).....(((((((...((((((....))))))........)))).))).(((.........))).)))).)))))........ ( -23.60) >consensus GCGGCUUUUGAGGGUUCUGAAAACAAAAUAUGGAAACCAUUAAACACCAGAAUCGUGGGGCAAAUAACAUUUAGCUGCUCCAUGUAAGCUAAAG__UUGCCACCGGCAAACGGAACUAAC ............(((((((.............................((...(((((((((.............)))))))))....))......(((((...))))).)))))))... (-21.06 = -21.00 + -0.06)

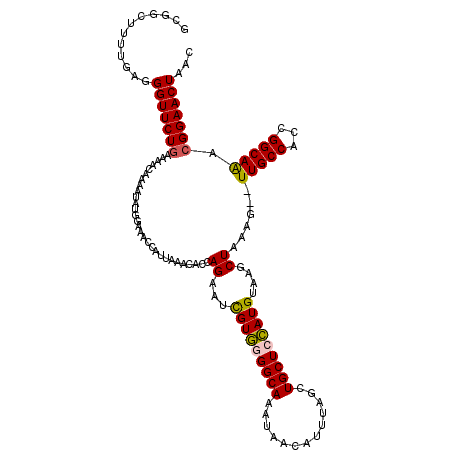

| Location | 7,768,396 – 7,768,489 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -24.47 |
| Energy contribution | -25.78 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7768396 93 - 22224390 AUGAUACAAACACAAGCGAACACAUUACAUGUACAUGUGUACA--------------------------UGCACGCAAGACCUGCAAAACAAGCAUACUUUUAAGCGCCAAAGUUAGGA ......(.(((....(((.........(((((((....)))))--------------------------))...(((.....)))....................)))....))).).. ( -14.50) >DroSec_CAF1 2500 119 - 1 AUGAUACAAACACAAGCGAACACAUUACAUGUACAUGUGUACGGUGGGCACAUCCUGUGCUGCCCACUGUGCACGCAUGACCUGCAAAACAAGCAUACUUUUAAGCGCCAAAGUUAGGA ......(.(((....(((.........(((((....(((((((((((((((((...))).)))))))))))))))))))...(((.......)))..........)))....))).).. ( -36.20) >DroSim_CAF1 2529 119 - 1 AUGAUACAAAAACAAGCGAACACAUUACAUGUACAUGUGUACAGUGGGCACAUCCUGUGCUGCCCACUGUGCACGCAAGACCUGCAAAACAAGCAUACUUUUAAGCGCCAAAGUUAGGA ...............(((..........((((....(((((((((((((((((...))).))))))))))))))(((.....))).......)))).........)))........... ( -35.51) >consensus AUGAUACAAACACAAGCGAACACAUUACAUGUACAUGUGUACAGUGGGCACAUCCUGUGCUGCCCACUGUGCACGCAAGACCUGCAAAACAAGCAUACUUUUAAGCGCCAAAGUUAGGA ...............(((..........((((....((((((((((((((((.....)).))))))))))))))(((.....))).......)))).........)))........... (-24.47 = -25.78 + 1.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:12 2006