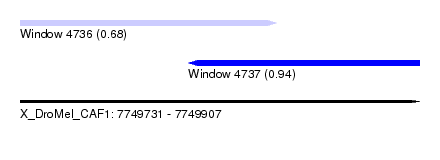
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,749,731 – 7,749,907 |
| Length | 176 |
| Max. P | 0.939433 |

| Location | 7,749,731 – 7,749,844 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -16.21 |
| Energy contribution | -18.35 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
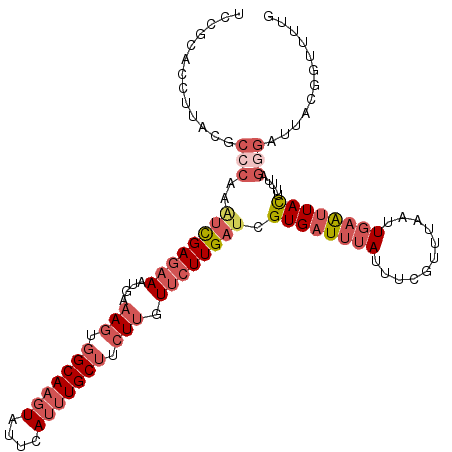
>X_DroMel_CAF1 7749731 113 + 22224390 UCCGCACCUUACGCCCAAUUCGAGAAAUGAAAGUGGCAAGUAUUCAUUUGCUUCUUGUUCUUGAUCGUGAUUUAUUUCGUUUAAUUGAAUUACUUUUAGGGAUUACGGUUUUG .....(((.((..(((...(((((((....(((.(((((((....))))))).))).)))))))..((((((((...........)))))))).....)))..)).))).... ( -28.30) >DroPse_CAF1 9516 91 + 1 UCCUUACCAUAGCUCUAAACUGA--AAAUAAAUCUGCACGUAUUCAAUUGCUUCUUGUUCUUGAUCGUGUUUUAUUUACUUUCACUUUGUUAU-------------------- ........(((((...(((.(((--((.(((((..(((((...((((..((.....))..)))).)))))...))))).))))).))))))))-------------------- ( -17.40) >DroSec_CAF1 9185 113 + 1 UCCGCACCUUACGCCCAAAUCGAGAAAUGAAAGUGGCAAGUAUUCAUUUGCUUCUUGUUCUUGAUCGUGAUUUAUUUCGUUUAAUUGAAUUACUUUUAGGGGUUACGGUUUUG .....(((..((.(((..((((((((....(((.(((((((....))))))).))).)))))))).((((((((...........)))))))).....)))))...))).... ( -29.60) >DroSim_CAF1 9349 113 + 1 UCCGCACCUUACGCCCAAAUCGAGAAAUGACAGUGGCAAGUAUUCAUUUGCUUCUUGUUCUUGAUCGUGAUUUAUUUCGUUUAAUUGAAUUACUUUUAGGGAUUACGGUUUUG .....(((.((..(((..((((((((.....((.(((((((....))))))).))..)))))))).((((((((...........)))))))).....)))..)).))).... ( -26.60) >DroEre_CAF1 10709 111 + 1 UCCGCACCUGACGCCCAAAUCGAGAAAUGAAAGUGGCAAGUAUUCAUUUGCUUCUUGUUCUUGAUCGUGAUUUAUUUCGUUUAAUUGAAUUACUUUUAGGGGUUUCGAUUU-- ........(((.((((..((((((((....(((.(((((((....))))))).))).)))))))).((((((((...........))))))))......)))).)))....-- ( -29.60) >DroYak_CAF1 10760 113 + 1 UCCUCACCUGACGCCCGAGUCGAGAAAUGAAAGUGGCAAGUAUUCAUUUGCUUCUUGUUCUUGAUCGUGAUUUAUUUCGUUUAAUUGAAUUACUUCAAGUGAUUUCGGUUUUG ...((((.(((.....((.(((((((....(((.(((((((....))))))).))).)))))))))((((((((...........)))))))).))).))))........... ( -27.50) >consensus UCCGCACCUUACGCCCAAAUCGAGAAAUGAAAGUGGCAAGUAUUCAUUUGCUUCUUGUUCUUGAUCGUGAUUUAUUUCGUUUAAUUGAAUUACUUUUAGGGAUUACGGUUUUG .............(((..((((((((....(((.(((((((....))))))).))).)))))))).((((((((...........)))))))).....)))............ (-16.21 = -18.35 + 2.14)

| Location | 7,749,805 – 7,749,907 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -15.37 |
| Consensus MFE | -12.94 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
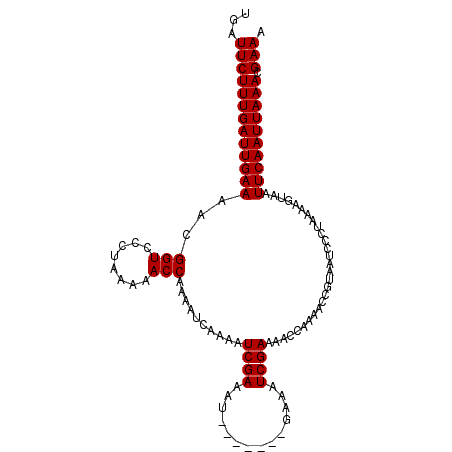
>X_DroMel_CAF1 7749805 102 - 22224390 UGAUUCUUUGAUUGAAAACGGUCCCUAAAAACCAAAAUCAAAAUCGAAAU------UGAAAUCGAAAACCAAAACCGUAAUCCCUAAAAGUAAUUCAAUUAAACGAAA ...(((((((((((((...(((........)))..........((((...------.....))))............................)))))))))).))). ( -14.30) >DroSec_CAF1 9259 96 - 1 UGAUUCUUUGAUUGAAAACGGUCCCUAAAAACCAAAAUCAAAAUCGA------------AAUCGAAAACCAAAACCGUAACCCCUAAAAGUAAUUCAAUUAAACGAAA ...(((((((((((((...(((........)))..........(((.------------...)))............................)))))))))).))). ( -13.70) >DroSim_CAF1 9423 108 - 1 UGAUUCUUUGAUUGAAAACGGUCCCUAAAAACCAAAAUCAAAAUCGAAAUCGAAAUCGAAAUCGAAAACCAAAACCGUAAUCCCUAAAAGUAAUUCAAUUAAACGAAA ...(((((((((((((...(((........)))..........((((..(((....)))..))))............................)))))))))).))). ( -18.10) >consensus UGAUUCUUUGAUUGAAAACGGUCCCUAAAAACCAAAAUCAAAAUCGAAAU_______GAAAUCGAAAACCAAAACCGUAAUCCCUAAAAGUAAUUCAAUUAAACGAAA ...(((((((((((((...(((........)))..........((((..............))))............................)))))))))).))). (-12.94 = -12.94 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:57 2006