
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,709,544 – 7,709,690 |
| Length | 146 |
| Max. P | 0.893158 |

| Location | 7,709,544 – 7,709,664 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -39.31 |
| Consensus MFE | -36.23 |
| Energy contribution | -37.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7709544 120 - 22224390 CCCGCCGCACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCCAUCGACUGGUUGGUUACAUGUAAGUGCUGCCAUUGCCCCC ......(((((((((((.......)))))))))))........((((((....(((.((((((((...((((..((((....))))....))))....)))))))).))).))))))... ( -40.90) >DroSim_CAF1 17394 120 - 1 CCCGCCGCACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCCAUCGACUGGUUGGUUACAUGUAAGUGCUGCCAUUGCCCCC ......(((((((((((.......)))))))))))........((((((....(((.((((((((...((((..((((....))))....))))....)))))))).))).))))))... ( -40.90) >DroEre_CAF1 7135 116 - 1 CCCGCCACACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCCAUCGACUG----GUUACAUGUAAGUGCUGCCAUUGCCCCC ...((.(((((((.............)))))))))........((((((....(((.((((((((...((((..((((....))))))----))....)))))))).))).))))))... ( -36.12) >consensus CCCGCCGCACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCCAUCGACUGGUUGGUUACAUGUAAGUGCUGCCAUUGCCCCC ......(((((((((((.......)))))))))))........((((((....(((.((((((((...((((..((((....))))....))))....)))))))).))).))))))... (-36.23 = -37.23 + 1.00)

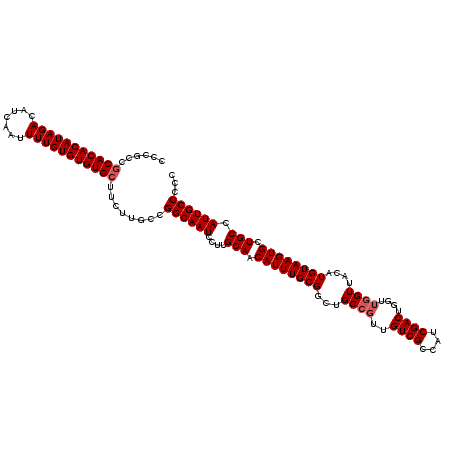
| Location | 7,709,584 – 7,709,690 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -22.97 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7709584 106 - 22224390 AAACC---GAA-CUCCGAAACCCC----------AAUCCACCCGCCGCACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCC .....---...-...(((......----------............(((((((((((.......))))))))))).......(((((..(...(((.....))))..)))))...))).. ( -25.50) >DroSim_CAF1 17434 106 - 1 AAACU---GAA-CUCCGAAACCCC----------AAUCCACCCGCCGCACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCC .....---...-...(((......----------............(((((((((((.......))))))))))).......(((((..(...(((.....))))..)))))...))).. ( -25.50) >DroEre_CAF1 7171 120 - 1 AAACCCCCAAACCCCCAAAACCCCCAAAAACCGCCCCCAACCCGCCACACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCC ................................(((..(((...((.(((((((.............)))))))))...)))..)))................(((((.......))))). ( -23.32) >consensus AAACC___GAA_CUCCGAAACCCC__________AAUCCACCCGCCGCACACAUAGACAUCAAUUUUGUGUGUGCUUCUUGCCGGCAAUCCUUGCAACAUUUGCGGCUGCCGUUGUCGCC ...........................................((.(((((((((((.......))))))))))).......(((((..(...(((.....))))..))))).....)). (-22.97 = -23.30 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:50 2006