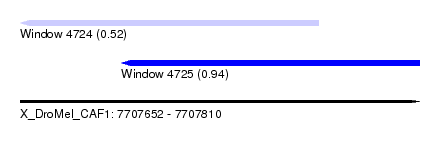
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,707,652 – 7,707,810 |
| Length | 158 |
| Max. P | 0.937078 |

| Location | 7,707,652 – 7,707,770 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -24.19 |
| Energy contribution | -25.82 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7707652 118 - 22224390 AUCCGGCCACGCCCCGUCCUGCCCACCUA--AACGCAAAGCCCACAAAGGCAGCGGGAACAACCGCUGCACAAUGUGGCAAAUGUUGGCCACUCAUUACGGAUUGCCGAGGUGCCAACUG .....((((((....((..(((.......--...)))..))........(((((((......)))))))....))))))....((((((.(((.....(((....))).))))))))).. ( -38.90) >DroSec_CAF1 6263 90 - 1 --------------------------CUA--AACGCAACGCCCAC--UGGCAGCGGGAACAACCGCCGCACAAUGUGGCAAAUGUUGGCCACUCAUUACGGAUUGCCGAGGUGCCAACUG --------------------------...--........(((((.--((((.((((......)))).)).)).)).)))....((((((.(((.....(((....))).))))))))).. ( -31.70) >DroSim_CAF1 15545 116 - 1 AUCCGGCCACGCCACGUCCUGCCCACCUA--AACGCAACGCCCAC--UGGCAGCGGGAACAACCGCCGCACAAUGUGGCAAAUGUUGGCCACUCAUUACGGAUUGCCGAGGUGCCAACUG ..........(((((((..(((.......--........(((...--.))).((((......)))).)))..)))))))....((((((.(((.....(((....))).))))))))).. ( -39.20) >DroEre_CAF1 5302 101 - 1 AUCCGGCCACGCCCCUUCCUGC----CCACUGGGGCA---------------GCGGUAGCAAGCGCCGCACAAUGUGGCAAAUGUUGGCCACUCAUUACGGAUUGUCGAGGUGCCAGCCG .....((((((((((.......----.....))))).---------------(((((.......))))).....)))))....((((((..(((((........)).)))..)))))).. ( -40.00) >consensus AUCCGGCCACGCCCCGUCCUGC____CUA__AACGCAACGCCCAC__UGGCAGCGGGAACAACCGCCGCACAAUGUGGCAAAUGUUGGCCACUCAUUACGGAUUGCCGAGGUGCCAACUG .....(((((((........))............((...(((......))).((((......)))).)).....)))))....((((((..(((.............)))..)))))).. (-24.19 = -25.82 + 1.63)

| Location | 7,707,692 – 7,707,810 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -32.98 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7707692 118 - 22224390 AGGAGGCGGCUUAGGACCAAGUUCGCUGCACAGCCGCCACAUCCGGCCACGCCCCGUCCUGCCCACCUA--AACGCAAAGCCCACAAAGGCAGCGGGAACAACCGCUGCACAAUGUGGCA ((((((((((((((((......)).)))...)))))))......(((...)))...)))).........--........(((.(((...(((((((......)))))))....)))))). ( -44.00) >DroSim_CAF1 15585 116 - 1 AGGAGGCGGCUUAGGACCAAGUUCGCUGCACAGCCGCCACAUCCGGCCACGCCACGUCCUGCCCACCUA--AACGCAACGCCCAC--UGGCAGCGGGAACAACCGCCGCACAAUGUGGCA .(((((((((((((((......)).)))...)))))))...)))......(((((((..(((.......--........(((...--.))).((((......)))).)))..))))))). ( -44.20) >DroEre_CAF1 5342 101 - 1 AGGACGCGGCUUAGGAGCAAGUUCGCUGCACAGCCGCCACAUCCGGCCACGCCCCUUCCUGC----CCACUGGGGCA---------------GCGGUAGCAAGCGCCGCACAAUGUGGCA .(((.((((((..(.(((......))).)..))))))....))).((((((((((.......----.....))))).---------------(((((.......))))).....))))). ( -46.50) >consensus AGGAGGCGGCUUAGGACCAAGUUCGCUGCACAGCCGCCACAUCCGGCCACGCCCCGUCCUGCCCACCUA__AACGCAA_GCCCAC___GGCAGCGGGAACAACCGCCGCACAAUGUGGCA .(((((((((((((((......)).)))...)))))))...))).(((((((........))............((...(((......))).((((......)))).)).....))))). (-32.98 = -33.90 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:46 2006