
| Sequence ID | X_DroMel_CAF1 |
|---|---|
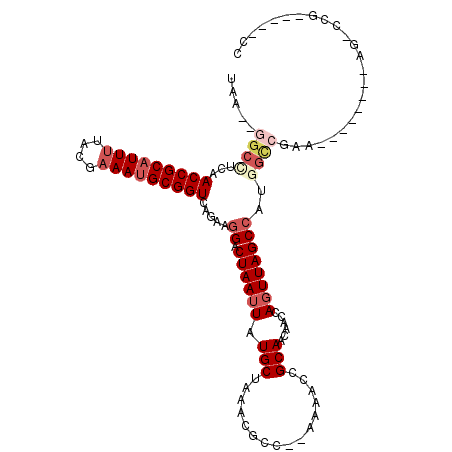
| Location | 7,689,954 – 7,690,053 |
| Length | 99 |
| Max. P | 0.957310 |

| Location | 7,689,954 – 7,690,053 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.96 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
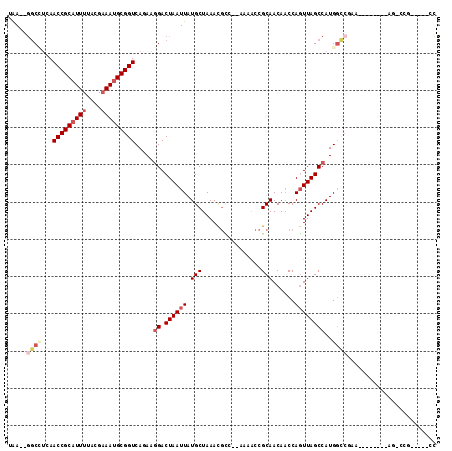
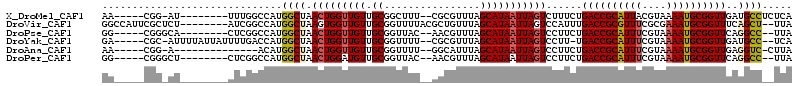
>X_DroMel_CAF1 7689954 99 + 22224390 UGAGAGGCAUCAACCGCAUUUUACGUAAUGCGGUCAGAAAGACUAAUUAUGCUAAACGCG--AAAGCCGCAACAACCAGUUAGCCAUGGCCAAA--------AU-CCG-----UU .....(((.((.((((((((......))))))))..))..((((.....(((.....((.--...)).)))......)))).)))((((.....--------..-)))-----). ( -23.50) >DroVir_CAF1 9880 105 + 1 UAA--AGCUGAAACCGCAUUUCGCGAAACGCGGUCAAAUGGACUAAUUAUGCUAAACAGCGUAAAACCGCAACAACCACUUAGCCAUGGCCGAU--------AGAGCGAAUGGCC ...--..........((..(((((......((((((..(((.(((((((((((....)))))))...............)))))))))))))..--------...)))))..)). ( -27.06) >DroPse_CAF1 29803 98 + 1 UAA--GGCCUGAACCGCAUUUUACGAAAUGCGGUCAGAAGGACUAAUUAUGCUAAACGUU--GUAACCGCAACAACCAGUUAGCCAUGGCCGAG--------UGCCCG-----CC ...--(((((((.((((((((....))))))))))))..(((((..(((((((((..(((--((.......)))))...)))).)))))...))--------).)).)-----)) ( -32.10) >DroYak_CAF1 7988 104 + 1 UGA--GGCAUCAACCGCAUUUUACGAAAUGCGGUCA-AAGGACUAAUUAUGCUAAACGCG--AAAACCGCAACAACCAGUUAGCCAUGGUCAAAAUAAUAAAAU-GCG-----UC .((--.((((..(((((((((....)))))))))..-...(((((.....(((....(((--.....)))(((.....))))))..)))))...........))-)).-----)) ( -25.90) >DroAna_CAF1 53093 92 + 1 UAAG-GACCUCAACCGCAUUUUACGAAAUGCGGUCAGAAGGACUAAUUAUGCUAAAUGCC--AAAACCGCAACAACCAGUUAGCCAUGU--------------U-CCG-----UU ...(-((.....(((((((((....))))))))).....((.((((((........(((.--......)))......))))))))....--------------)-)).-----.. ( -20.64) >DroPer_CAF1 30374 98 + 1 UAA--GGCCUGAACCGCAUUUUACGAAAUGCGGUCAGAAGGACUAAUUAUGCUAAACGUU--GUAACCGCAACAUCCAGUUAGCCAUGGCCGAG--------AGCCCG-----CC ...--(((((((.((((((((....))))))))))))..((.((((((..(.....)(((--((....)))))....))))))))..((.(...--------.).)))-----)) ( -30.20) >consensus UAA__GGCCUCAACCGCAUUUUACGAAAUGCGGUCAGAAGGACUAAUUAUGCUAAACGCC__AAAACCGCAACAACCAGUUAGCCAUGGCCGAA________AG_CCG_____CC .....((((...(((((((((....))))))))).....((.((((((.(((................)))......))))))))..))))........................ (-15.48 = -16.96 + 1.47)

| Location | 7,689,954 – 7,690,053 |
|---|---|
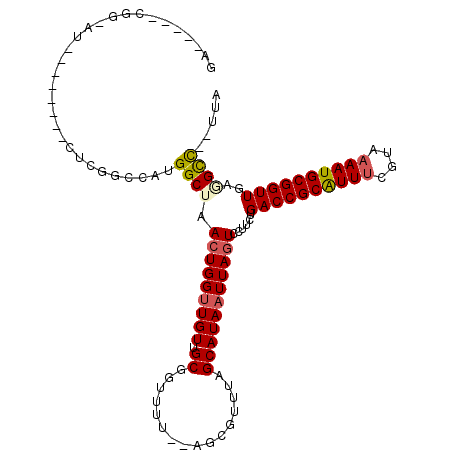
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7689954 99 - 22224390 AA-----CGG-AU--------UUUGGCCAUGGCUAACUGGUUGUUGCGGCUUU--CGCGUUUAGCAUAAUUAGUCUUUCUGACCGCAUUACGUAAAAUGCGGUUGAUGCCUCUCA ..-----...-..--------...(((...((((((...((((.((((.....--))))..))))....)))))).....(((((((((......)))))))))...)))..... ( -27.70) >DroVir_CAF1 9880 105 - 1 GGCCAUUCGCUCU--------AUCGGCCAUGGCUAAGUGGUUGUUGCGGUUUUACGCUGUUUAGCAUAAUUAGUCCAUUUGACCGCGUUUCGCGAAAUGCGGUUUCAGCU--UUA ((((.........--------...))))((((((((...((((..(((......)))....))))....)))).))))..((((((((((....))))))))))......--... ( -32.80) >DroPse_CAF1 29803 98 - 1 GG-----CGGGCA--------CUCGGCCAUGGCUAACUGGUUGUUGCGGUUAC--AACGUUUAGCAUAAUUAGUCCUUCUGACCGCAUUUCGUAAAAUGCGGUUCAGGCC--UUA ((-----(((((.--------...(((....)))..((((.(((((......)--)))).))))........))))..((((((((((((....)))))))).)))))))--... ( -34.60) >DroYak_CAF1 7988 104 - 1 GA-----CGC-AUUUUAUUAUUUUGACCAUGGCUAACUGGUUGUUGCGGUUUU--CGCGUUUAGCAUAAUUAGUCCUU-UGACCGCAUUUCGUAAAAUGCGGUUGAUGCC--UCA ((-----.((-(((................((((((...((((.((((.....--))))..))))....))))))...-.((((((((((....))))))))))))))).--)). ( -29.70) >DroAna_CAF1 53093 92 - 1 AA-----CGG-A--------------ACAUGGCUAACUGGUUGUUGCGGUUUU--GGCAUUUAGCAUAAUUAGUCCUUCUGACCGCAUUUCGUAAAAUGCGGUUGAGGUC-CUUA ..-----.((-(--------------....((((((...((((.(((......--.)))..))))....))))))((((.((((((((((....))))))))))))))))-)... ( -28.30) >DroPer_CAF1 30374 98 - 1 GG-----CGGGCU--------CUCGGCCAUGGCUAACUGGAUGUUGCGGUUAC--AACGUUUAGCAUAAUUAGUCCUUCUGACCGCAUUUCGUAAAAUGCGGUUCAGGCC--UUA ((-----((((((--------...(((....)))..((((((((((......)--))))))))).......)))))..((((((((((((....)))))))).)))))))--... ( -39.00) >consensus GA_____CGG_AU________CUCGGCCAUGGCUAACUGGUUGUUGCGGUUUU__AGCGUUUAGCAUAAUUAGUCCUUCUGACCGCAUUUCGUAAAAUGCGGUUGAGGCC__UUA ..............................((((.(((((((((.((................)))))))))))......((((((((((....))))))))))..))))..... (-21.24 = -21.57 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:35 2006