
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,674,157 – 7,674,310 |
| Length | 153 |
| Max. P | 0.657313 |

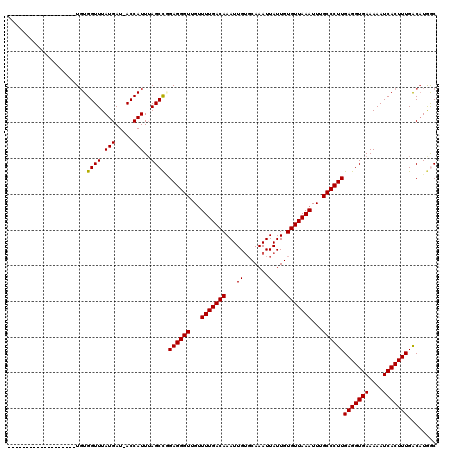
| Location | 7,674,157 – 7,674,257 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -23.00 |
| Energy contribution | -22.87 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7674157 100 + 22224390 -------------------UGUGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGAC -------------------..(((((.(((..-..)))..)))))((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))........ ( -24.70) >DroVir_CAF1 166601 111 + 1 --------CUGCCUGCUGUUGUGGUUUAUGAU-ACCAUUUAGCCCGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC --------..(((.((((..(((((.......-))))).)))).(((((((...(((((((....((....)).....)))))))...)))))))((((((....))))))......))) ( -31.90) >DroGri_CAF1 183650 119 + 1 UUUGCCUUCUGCUUGCUGUUGUGGUUUAUGAU-ACCAUUAAGCCCGAGGGUUGGUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC ...(((........(((...(((((.......-)))))..))).(((((((..((((((((....((....)).....))))))))..)))))))((((((....))))))......))) ( -34.20) >DroSec_CAF1 88385 100 + 1 -------------------UGUGGUUUAUGAU-ACCAUUUAGCUGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC -------------------.(((((((.((..-..))((((.((.((((((...(((((((....((....)).....)))))))...)))))).)).)))))))))))........... ( -25.40) >DroWil_CAF1 111657 111 + 1 --------CUCUCU-CCCUUUUGGUUUAUGAUGACCAUUUAGCCAGAGGGUUGUUUUGACAAAUUGGCCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGGCCUAGC --------......-.((((((((((.(((.....)))..))))))))))((((....))))...(((((((.....(((.....((..((.....))..))....)))))))))).... ( -29.80) >DroYak_CAF1 89117 100 + 1 -------------------UGUGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGAC -------------------..(((((.(((..-..)))..)))))((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))........ ( -24.70) >consensus ___________________UGUGGUUUAUGAU_ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC ......................((((.(((.....)))..)))).((((((...(((((((...((.......))...)))))))...))))))(((((((....)))))))........ (-23.00 = -22.87 + -0.14)


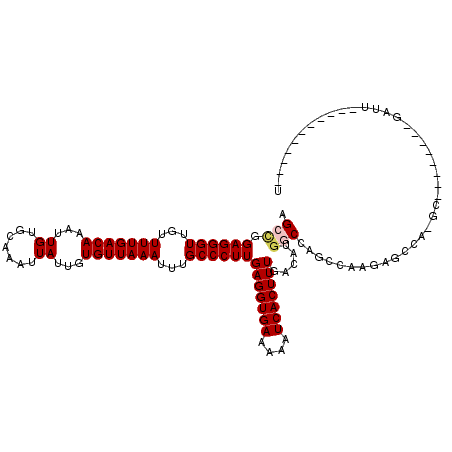
| Location | 7,674,177 – 7,674,277 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7674177 100 + 22224390 AGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGACCAGCCAAGAGGCA-GC--------GAUU-----------U .((.((.((((...(((((((....((....)).....)))))))...))))(..((((((....))))))..).....)).(((....))).-))--------....-----------. ( -27.20) >DroVir_CAF1 166632 96 + 1 AGCCCGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCG----CAAUGCCA-UG--------GAUU-----------A .((((((((((...(((((((....((....)).....)))))))...))))))).)))...............((((((.----....))))-))--------....-----------. ( -30.10) >DroGri_CAF1 183689 96 + 1 AGCCCGAGGGUUGGUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCG----CAAUGCCA-UG--------GAUU-----------A .((((((((((..((((((((....((....)).....))))))))..))))))).)))...............((((((.----....))))-))--------....-----------. ( -32.60) >DroSec_CAF1 88405 99 + 1 AGCUGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCA-GC--------GAUU------------ .((((((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))....(((....))).....))-))--------....------------ ( -28.80) >DroWil_CAF1 111688 119 + 1 AGCCAGAGGGUUGUUUUGACAAAUUGGCCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGGCCUAGCCAGCAAA-AGGCAAGUGGCAAGUGGAUUUCAUUUGGCUAU ((((.((((((...(((((((...(((.....)))...)))))))...)))))).((((((((..(((((...(((..(((......-.)))....))).))))).)))))))))))).. ( -37.70) >DroYak_CAF1 89137 100 + 1 AGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGACCAGCCAAGAGCCA-GC--------GAUU-----------U .((.((..(((((((.((.(((((((..(((....)))..)..))))))...(..((((((....))))))..))).)).))))).....)).-))--------....-----------. ( -23.60) >consensus AGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGCCA_GC________GAUU___________U .(((.((((((...(((((((...((.......))...)))))))...))))))(((((((....))))))).....)))........................................ (-20.39 = -20.78 + 0.39)

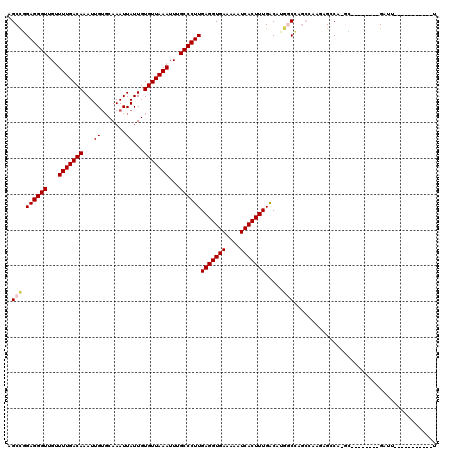
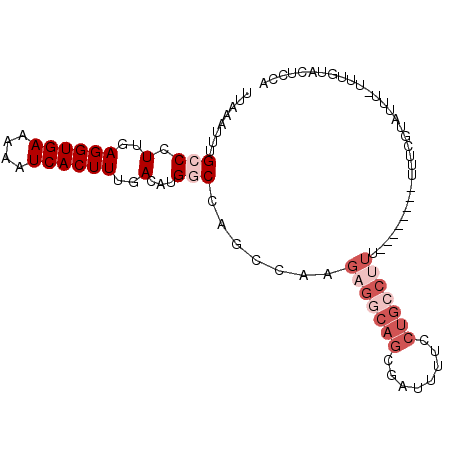
| Location | 7,674,217 – 7,674,310 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -11.53 |
| Energy contribution | -13.37 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7674217 93 + 22224390 UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGACCAGCCAAGAGGCAGCGAUUUUCCUGCCUUUU------UUUUCGUAUUU-UUUGUACUCCA ............(..((((((....))))))..)...........(((((((((.(.....))))))))))------.....((((..-...)))).... ( -20.40) >DroGri_CAF1 183729 70 + 1 UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCG----CAAUGCCAUGGAUUAUAUU-----------------UUUAUUU-UUU-------- ............(..((((((....))))))..)((((((.----....)))))).........-----------------.......-...-------- ( -16.40) >DroSec_CAF1 88445 90 + 1 UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAGCGAUU-UCCUGCCUUU--------UUUCGUAUUU-UUUGUACUCCA ........(((.(..((((((....))))))..)...)))..(..(((((((((.(...-.)))))))))--------)..)((((..-...)))).... ( -24.50) >DroSim_CAF1 67101 92 + 1 UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAGCGAUUUUCCUGCCUUU--------UUUCGUAUUUUUUUGUACUCCA ........(((.(..((((((....))))))..)...)))..(..(((((((((.(.....)))))))))--------)..)((((......)))).... ( -24.70) >DroEre_CAF1 87637 95 + 1 UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAGCGAUUUUCCUGCCCUUU----UUCCCUCGUAUUU-UUUGCACUCCA ..............((((.((((((...........(((....)))...(((((.(.....)))))).)))----)))))))(((...-..)))...... ( -22.30) >DroYak_CAF1 89177 99 + 1 UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGACCAGCCAAGAGCCAGCGAUUUUCCAGCCUUCUCGCAUUUUCUCGCAUUU-UCCGUACUCCA .......(((..(..((((((....))))))..).............(((...((((.............)))).....))))))...-........... ( -14.22) >consensus UUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAGCGAUUUUCCUGCCUUU________UUUCGUAUUU_UUUGUACUCCA ........(((.(..((((((....))))))..)...))).......(((((((........)))))))............................... (-11.53 = -13.37 + 1.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:27 2006