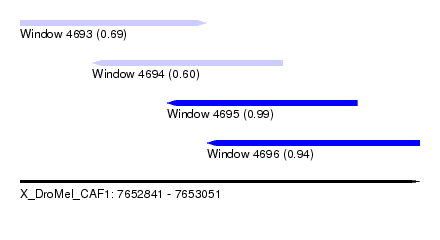
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,652,841 – 7,653,051 |
| Length | 210 |
| Max. P | 0.991063 |

| Location | 7,652,841 – 7,652,939 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.66 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7652841 98 + 22224390 UCGC--CACACACUUUUCACCUGCUGCAAGAUUUUCUUCUGCUCUGCUUUUCUCCGUUCUCUUGGCUGGCAAAUUGCCCAGCAACAACAACAGCCUCAAU ..((--................((.(((.((......)))))...))........(((......((((((.....).)))))......))).))...... ( -15.60) >DroSec_CAF1 69054 98 + 1 CCGA--UGCACACUUUUCACCUGCUGCACGAUUUUCUUCUGCUCUGCUUUUCUCUGUUCUCUUGGCUGGCAAAUUGCCCAGCAACAACAACAGCCUCAAU ..((--.((.............((.(((.((......)))))...)).......((((......((((((.....).)))))......)))))).))... ( -19.20) >DroSim_CAF1 56951 98 + 1 CCGC--UGCACACUUUUCACCUGCUGCAAGAUUUUCUUCUGCUCUGCUUUUCUCUGUUCUCUUGGCUGGCAAAUUGCCCAGCAACAACAACAGCCUCAAU ..((--((..............((.(((.((......)))))...)).......((((......((((((.....).)))))...)))).))))...... ( -19.00) >DroEre_CAF1 74863 100 + 1 CCACCCCUCACACUUUCCACCUGCUGCAAGAUUUUCUUAUGCUCUGCUUUUCCCUGUUCUCUUGGCUGGCAAAUUGCCCAGCAACAAAAACAGCCUCAAU ......................((.(((((.....))..)))...))......(((((......((((((.....).)))))......)))))....... ( -16.00) >DroYak_CAF1 74714 98 + 1 UCAC--CACACACUUUUCACCUGCUGCAAGAUUUUCCUCGGCUCCGCUUUUCUCCGUUCUCCUGGCUGGCAAAUUGCCCAGCAACAAAAACAGCCUCAAU ....--................((((............((....))..................((((((.....).)))))........))))...... ( -13.30) >consensus CCGC__CACACACUUUUCACCUGCUGCAAGAUUUUCUUCUGCUCUGCUUUUCUCUGUUCUCUUGGCUGGCAAAUUGCCCAGCAACAACAACAGCCUCAAU ......................(((((((((.........((...))............)))))((((((.....).)))))........))))...... (-13.26 = -13.66 + 0.40)


| Location | 7,652,879 – 7,652,979 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -22.86 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7652879 100 - 22224390 GACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGA-------------------GGCUGUUGUUGUUGCUGGGCAAUUUGCCAGC-CAAGAGAACGGAGAAAAGCAGAGC ..(((.....)))((..((((((((((((.....)))))))))))-------------------).(((((.((...(((((........)))))-...)).)))))......))..... ( -28.90) >DroSec_CAF1 69092 100 - 1 GACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGA-------------------GGCUGUUGUUGUUGCUGGGCAAUUUGCCAGC-CAAGAGAACAGAGAAAAGCAGAGC ..(((.....)))((..((((((((((((.....)))))))))))-------------------).(((((.((...(((((........)))))-...)).)))))......))..... ( -29.60) >DroSim_CAF1 56989 100 - 1 GACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGA-------------------GGCUGUUGUUGUUGCUGGGCAAUUUGCCAGC-CAAGAGAACAGAGAAAAGCAGAGC ..(((.....)))((..((((((((((((.....)))))))))))-------------------).(((((.((...(((((........)))))-...)).)))))......))..... ( -29.60) >DroEre_CAF1 74903 100 - 1 GACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGA-------------------GGCUGUUUUUGUUGCUGGGCAAUUUGCCAGC-CAAGAGAACAGGGAAAAGCAGAGC ..(((.....)))((..((((((((((((.....)))))))))))-------------------).((((((((...(((((........)))))-...))))))))......))..... ( -31.30) >DroYak_CAF1 74752 100 - 1 GACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGA-------------------GGCUGUUUUUGUUGCUGGGCAAUUUGCCAGC-CAGGAGAACGGAGAAAAGCGGAGC ..(((.....)))((..((((((((((((.....)))))))))))-------------------).((((((((.(.(((((........)))))-.).))))))))......))..... ( -31.30) >DroPer_CAF1 97553 114 - 1 AGUAGACUGGCUGGCAAUUCAAU-UGCUAACCAGUGGAAAAUUGCUAGCAUUUACACUGCUGUCUGCUGUCUUUGUUGUUGGGCAAUUUGCCAGAAUCAGCGAGCAG---AAAGCACA-- .(((((...((((((((((...(-..((....))..)..)))))))))).)))))..((((.((((((.....(((((...(((.....))).....))))))))))---).))))..-- ( -40.10) >consensus GACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGA___________________GGCUGUUGUUGUUGCUGGGCAAUUUGCCAGC_CAAGAGAACAGAGAAAAGCAGAGC ..(((.....)))((...(((((((((((.....))))))))))).....................(((((.((...(((((........)))))....)).)))))......))..... (-22.86 = -23.08 + 0.22)

| Location | 7,652,918 – 7,653,018 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 95.80 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.26 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7652918 100 - 22224390 AGUCGUUGUUUUUGCCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGAGGCUGUUGUUGUUGCU ((((.......(((((.(.(((....))))......((((((...))))))))))).(((((((((((.....)))))))))))))))............ ( -27.90) >DroSec_CAF1 69131 100 - 1 AGUCGUUGUUUUUGUCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGAGGCUGUUGUUGUUGCU .((((.((.((((((((.((((...........)))).)))))))).)).))))..((((((((((((.....))))))))))))((..........)). ( -28.40) >DroSim_CAF1 57028 100 - 1 AGUCGUUGUUUUUGUCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGAGGCUGUUGUUGUUGCU .((((.((.((((((((.((((...........)))).)))))))).)).))))..((((((((((((.....))))))))))))((..........)). ( -28.40) >DroEre_CAF1 74942 100 - 1 AGUCAGUGUUUUUGUCAGUGUGAAAGGAGCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGAGGCUGUUUUUGUUGCU .(((((((.((((((((.(((.............))).)))))))).)))))))..((((((((((((.....))))))))))))((..........)). ( -31.72) >DroYak_CAF1 74791 100 - 1 AGGCGUUGUUUUUGUCAGUGUGAAAAUAGCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGAGGCUGUUUUUGUUGCU .((((.......))))((((..((((((((...(.(((((((...))))))).)..((((((((((((.....))))))))))))))))))))..).))) ( -30.40) >consensus AGUCGUUGUUUUUGUCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAAAUUGAGGCUGUUGUUGUUGCU .((((.((.((((((((.((((...........)))).)))))))).)).))))..((((((((((((.....))))))))))))((..........)). (-24.62 = -25.26 + 0.64)


| Location | 7,652,939 – 7,653,051 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7652939 112 - 22224390 AACAUUUA---UCCAGAUUUAAAUGGUAGAGA--UUUUAGUCGUUGUUUUUGCCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAA ..((((((---........)))))).......--.....(((((((((((((.....(((....))))))))))))))))...(((((((((((......)))....)))))))).. ( -28.30) >DroSec_CAF1 69152 112 - 1 CAUAUUUA---AACAGACUUAAAUGAUAGAGA--AUUUAGUCGUUGUUUUUGUCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAA ........---(((.((((.((((........--)))))))))))...(((((((.((((...........)))).)))))))(((((((((((......)))....)))))))).. ( -29.20) >DroSim_CAF1 57049 112 - 1 CAUAUUUA---AACAGACUUAAACGAUAGAGA--CUUUAGUCGUUGUUUUUGUCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAA ........---..........((((((...((--(....)))))))))(((((((.((((...........)))).)))))))(((((((((((......)))....)))))))).. ( -28.20) >DroEre_CAF1 74963 116 - 1 CAUACAUACGACACAGACUGGAAAACUAGAGAG-AUUUAGUCAGUGUUUUUGUCAGUGUGAAAGGAGCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAA ................(((((..(((.(((..(-(.((.(((((((.((((((((.(((.............))).)))))))).))))))))).))..))))))..)))))..... ( -30.52) >DroYak_CAF1 74812 105 - 1 ------------AUAAACUGAAAUCGUUGAGAGUAUUAAGGCGUUGUUUUUGUCAGUGUGAAAAUAGCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAA ------------.....(((....(.((((.....)))).)((((((((((((...(((....))))))))))))))).))).(((((((((((......)))....)))))))).. ( -24.00) >consensus CAUAUUUA___AACAGACUUAAAUGAUAGAGA__AUUUAGUCGUUGUUUUUGUCAGUGUCAAAAGACCAAAGAUAGUGACAGAGUCACUGGCAAUUCAAUUUGUUAACCAGUGGCAA .......................................(((((((((((((...............)))))))))))))...(((((((((((......)))....)))))))).. (-21.80 = -22.04 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:20 2006