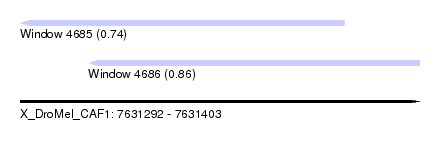
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,631,292 – 7,631,403 |
| Length | 111 |
| Max. P | 0.863796 |

| Location | 7,631,292 – 7,631,382 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -12.75 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
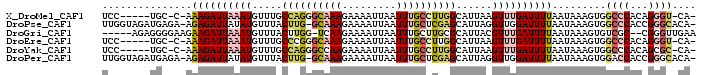
>X_DroMel_CAF1 7631292 90 - 22224390 UCC-----UGC-C-AAAGAUUAAAUGUUUGCCAGGGCAAAGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAUAAAGUGGCCCACAGGGU-CA- ...-----...-.-.((((((((((...((((((((((((.........))))))))))))....)))))))))).......((((((...))))-))- ( -33.20) >DroPse_CAF1 81151 96 - 1 UUGGUAGAUGAGA-AGAGAUUAUAUGUUUACUUG-GCAAAGAAAAUUAAUUUGCUCGAGCAUUAGGUUGGAUUUUAAUAAAGUGGCCCACCGGGCACA- ........(((((-...((((..((((((....(-(((((.........)))))).))))))..))))...))))).....(((.((....)).))).- ( -16.70) >DroGri_CAF1 137545 91 - 1 -----AGAGGGGAAGAAGAUUAAAUGUUUACUUGG-UCAAGAAAAUUAAUUUGCUUGCGCAUUACGUUUGAUUUUAAUAAAGUGUCGC--CGGGUUGAA -----...((.((..((((((((((((.....(((-.((((.(((....))).))))).))..)))))))))))).........)).)--)........ ( -15.50) >DroEre_CAF1 53906 90 - 1 UCC-----UGC-C-AAAGAUUAAAUGUUUGCCCGGGCAAAGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAUAAAGUGGCCCACAGGGU-CA- ...-----...-.-.((((((((((...((((.(((((((.........))))))).))))....)))))))))).......((((((...))))-))- ( -29.40) >DroYak_CAF1 51057 90 - 1 UCC-----UGC-C-AAAGAUUAAAUGUUUGCCAGGGCCAAGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAUAAAGUGGCCCACAGCGC-CA- ...-----.((-(-(((((((((((...(((((((((.((.........)).)))))))))....)))))))))).......)))).........-..- ( -25.91) >DroPer_CAF1 65293 96 - 1 UUGGUAGAUGAGA-AGAGAUUAUAUGUUUACUUG-GCAAAGAAAAUUAAUUUGCUCGAGCAUUAGGUUGGAUUUUAAUAAAGUGGACCACCGGGCACA- (((((.((((...-.(((.......((((.(((.-...))).)))).......)))...)))).((((.(.(((....))).).))))))))).....- ( -16.14) >consensus UCC_____UGC_A_AAAGAUUAAAUGUUUACCUGGGCAAAGAAAAUUAAUUUGCCUGGGCAUUAAGUUUGAUUUUAAUAAAGUGGCCCACAGGGC_CA_ ...............((((((((((.....((((((((((.........))))))))))......)))))))))).........((((...)))).... (-12.75 = -13.75 + 1.00)


| Location | 7,631,311 – 7,631,403 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7631311 92 - 22224390 UGUCCAGGGGCCUAAAACUAUUCC-----UGC-CAAAGAUUAAAUGUUUGCCAGGGCAA-AGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAU ....((((((..........))))-----)).-..((((((((((...(((((((((((-(.........))))))))))))....))))))))))... ( -30.30) >DroPse_CAF1 81171 97 - 1 GGACCCCCAUCUCGAAACUAUUUGGUAGAUGAGAAGAGAUUAUAUGUUUACUUG-GCAA-AGAAAAUUAAUUUGCUCGAGCAUUAGGUUGGAUUUUAAU .((((.....(((((........(((((((...............)))))))..-((((-(.........)))))))))).....)))).......... ( -15.66) >DroSim_CAF1 49955 93 - 1 UGUCCAGGGGCCUAAAACUAUUCC-----UGC-CAAAGAUUAAAUGUUUGCCAGGCCAAAAGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAU ....((((((..........))))-----)).-..((((((((((...(((((((.((((..........)))).)))))))....))))))))))... ( -22.30) >DroEre_CAF1 53925 92 - 1 UGUCCAGAGGGCCAAAACUAUUCC-----UGC-CAAAGAUUAAAUGUUUGCCCGGGCAA-AGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAU .........(((............-----.))-).((((((((((...((((.((((((-(.........))))))).))))....))))))))))... ( -25.12) >DroYak_CAF1 51076 92 - 1 UGUCCAGGGGCCUAAAACUAUUCC-----UGC-CAAAGAUUAAAUGUUUGCCAGGGCCA-AGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAU ....((((((..........))))-----)).-..((((((((((...(((((((((.(-(.........)).)))))))))....))))))))))... ( -26.20) >DroPer_CAF1 65313 97 - 1 GGACCCCCAUCUCGAAACUAUUUGGUAGAUGAGAAGAGAUUAUAUGUUUACUUG-GCAA-AGAAAAUUAAUUUGCUCGAGCAUUAGGUUGGAUUUUAAU .((((.....(((((........(((((((...............)))))))..-((((-(.........)))))))))).....)))).......... ( -15.66) >consensus UGUCCAGGGGCCCAAAACUAUUCC_____UGC_CAAAGAUUAAAUGUUUGCCAGGGCAA_AGAAAAUUAAUUUGCCUUGGCAUUAAGUUUGAUUUUAAU ...................................((((((((((...(((((((((((............)))))))))))....))))))))))... (-16.37 = -16.98 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:11 2006