
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,618,902 – 7,618,999 |
| Length | 97 |
| Max. P | 0.633392 |

| Location | 7,618,902 – 7,618,999 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -16.16 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7618902 97 + 22224390 GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUCUCAAGAAAGCUGUCACACAGUCUGCCU-CCU-ACCUCCAU--------C--ACCCACCCCCCU----CGUUU ...............((((....((((((.....)))))).........((((.....))))))))..-...-........--------.--............----..... ( -15.00) >DroSec_CAF1 35968 95 + 1 GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUCUCAAGAAAGCUGUCACACAGUCUGCCU-CCU-CCCCACAU--------C--ACCCACCCCCCU------UUU ...............((((....((((((.....)))))).........((((.....))))))))..-...-........--------.--............------... ( -15.00) >DroSim_CAF1 37940 95 + 1 GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUUUCAAGAAAGCUGUCACACAGUCUGCCU-CCU-CCCUCCAU--------C--ACCCACCCCCCU------UUU ...............((((..((((((((.....)))))))).......((((.....))))))))..-...-........--------.--............------... ( -16.80) >DroEre_CAF1 41673 89 + 1 GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUCUCAAGAAAGCUGUCACACAGCCUCCCU-CCC-CCCGCCUU----UCAG---ACUCG---C------------ ...............((((((..((((((.....)))))).........((((.....))))......-...-........----...)---))).)---)------------ ( -18.20) >DroYak_CAF1 38218 101 + 1 GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUCUCAAGAAAGCUGUCACACAGUCUCCCU-CCU-UCCUCCUU----UCAUCUUACUCCUUCCCUC------UUU .............(((.(((...((((((.....)))))).....((((((((.....))).......-...-.....)))----)).....))).))).....------... ( -11.96) >DroAna_CAF1 34777 110 + 1 GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUCUCAAGAAAGCUGUCACACAGUUUGCCCUCCAACCCUCCAACCCUGCAG---CCCCACACCCCAGUGUCCUUU ...............((((....((((((.....)))))).......((((((.....))))))...................))))..---....((((....))))..... ( -20.00) >consensus GCAAAAUUUAUUUAAGCAGUCAAAUGCACGUGAAGUGCAUCUCAAGAAAGCUGUCACACAGUCUGCCU_CCU_CCCUCCAU________C__ACCCACCCCCCU______UUU ...............((((....((((((.....)))))).........((((.....))))))))............................................... (-13.16 = -13.22 + 0.06)

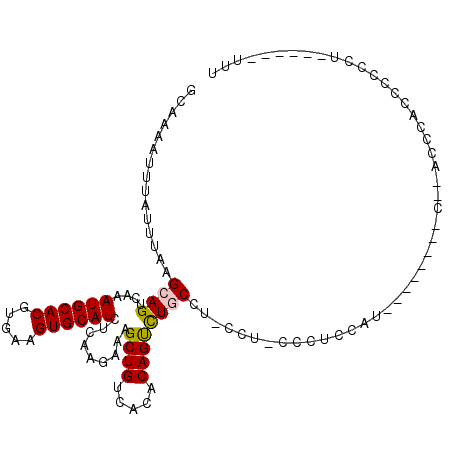
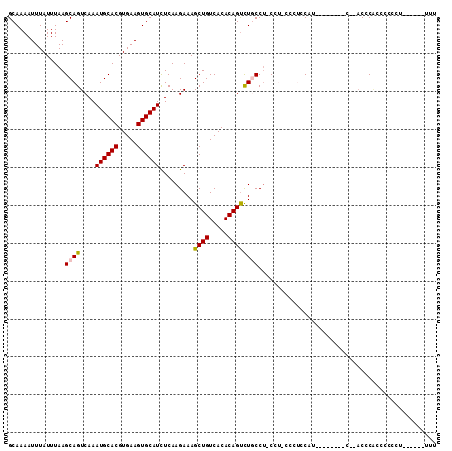
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:07 2006