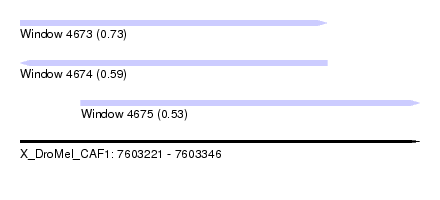
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,603,221 – 7,603,346 |
| Length | 125 |
| Max. P | 0.729175 |

| Location | 7,603,221 – 7,603,317 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
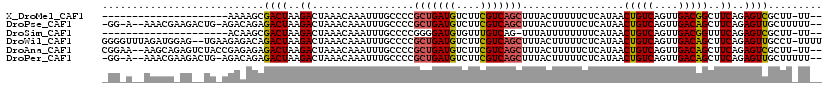
>X_DroMel_CAF1 7603221 96 + 22224390 ---------------------AAAAGCGACUAAGACUAAACAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUU-UU-- ---------------------((((((((((..((.................(((((((....))))))).................(((((....)))))..))..))))))))-))-- ( -29.20) >DroPse_CAF1 27116 113 + 1 -GG-A--AAACGAAGACUG-AGACAGAGACUAAGACUAAACAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCUUUUU-- -.(-(--((((((...(((-((...((((..(((......((....))....(((((((....)))))))....)))..))))....(((((....))))).)))))..))).)))))-- ( -27.10) >DroSim_CAF1 21061 95 + 1 ---------------------ACAAGCGACUAAGACUAAACAAAUUUGCCCCGGGGAUGUGUUUGUCAG-UUUAUUUUUUUUCAUAACUGUCAGUUGACGGUUUCAGAGUCGCUU-UU-- ---------------------..((((((((.(((((..((((((...((....))....)))))).))-)))............(((((((....)))))))....))))))))-..-- ( -28.00) >DroWil_CAF1 27473 117 + 1 GGGGUUUAGAUGGAG--UGAAGAGACAGACUAAGACUAAACAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCCU-UUUU (((((..(..((.((--(....((.....))...)))...))..)..)))))(((((((....)))))))..........(((....(((((....))))).....)))......-.... ( -28.80) >DroAna_CAF1 19388 115 + 1 CGGAA--AAGCAGAGUCUACCGAGAGAGACUAAGACUAAACAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUCGCUU-UU-- ...((--((((.((.(((...((((((((.((((......((....))....(((((((....))))))))))).))))))))....(((((....)))))....))).))))))-))-- ( -33.30) >DroPer_CAF1 26996 113 + 1 -GG-A--AAACGAAGACUG-AGACAGAGACUAAGACUAAACAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCUUUUU-- -.(-(--((((((...(((-((...((((..(((......((....))....(((((((....)))))))....)))..))))....(((((....))))).)))))..))).)))))-- ( -27.10) >consensus _GG_A__AAACG_AG__UG_AGAAAGAGACUAAGACUAAACAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUCGCUU_UU__ ...........................((((..((.................(((((((....))))))).................(((((....)))))..))..))))......... (-18.44 = -18.33 + -0.11)
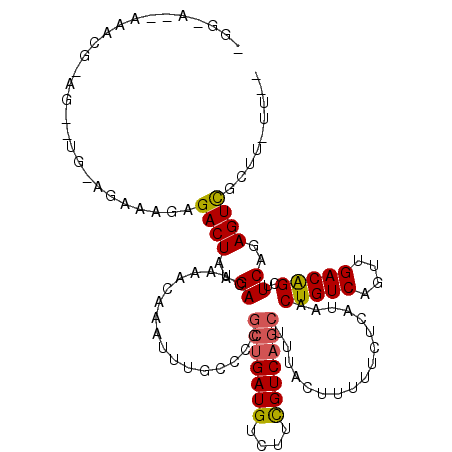
| Location | 7,603,221 – 7,603,317 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -18.01 |
| Energy contribution | -18.65 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7603221 96 - 22224390 --AA-AAGCGACUCUGAAGCCGUCAACUGACAGUUAUGAGAAAAAGUAAAGCUGACGAAGACAUCAGCGGGGCAAAUUUGUUUAGUCUUAGUCGCUUUU--------------------- --((-((((((((..(((((.(((....))).)))..........((...(((((........)))))...))............))..))))))))))--------------------- ( -26.60) >DroPse_CAF1 27116 113 - 1 --AAAAAGCAACUCUGAAGCUGUCAACUGACAGUUAUGAGAAAAAGUAAAGCUGACGAAGACAUCAGCGGGGCAAAUUUGUUUAGUCUUAGUCUCUGUCU-CAGUCUUCGUUU--U-CC- --.........(((...(((((((....)))))))..)))..........(..(((((((((....(((((((.................))).))))..-..))))))))).--.-).- ( -28.63) >DroSim_CAF1 21061 95 - 1 --AA-AAGCGACUCUGAAACCGUCAACUGACAGUUAUGAAAAAAAAUAAA-CUGACAAACACAUCCCCGGGGCAAAUUUGUUUAGUCUUAGUCGCUUGU--------------------- --..-((((((((..(((((.(((....))).)))...............-((((((((.....((....))....)))).))))))..))))))))..--------------------- ( -22.20) >DroWil_CAF1 27473 117 - 1 AAAA-AGGCAACUCUGAAGCUGUCAACUGACAGUUAUGAGAAAAAGUAAAGCUGACGAAGACAUCAGCGGGGCAAAUUUGUUUAGUCUUAGUCUGUCUCUUCA--CUCCAUCUAAACCCC ....-(((((.(((...(((((((....)))))))..))).............(((.(((((...((((((.....))))))..))))).)))))))).....--............... ( -25.90) >DroAna_CAF1 19388 115 - 1 --AA-AAGCGACUCUGAAGCUGUCAACUGACAGUUAUGAGAAAAAGUAAAGCUGACGAAGACAUCAGCGGGGCAAAUUUGUUUAGUCUUAGUCUCUCUCGGUAGACUCUGCUU--UUCCG --((-((((((.((((.(((((((....))))))).(((((.........(((((........)))))(((((.................)))))))))).)))).)).))))--))... ( -31.73) >DroPer_CAF1 26996 113 - 1 --AAAAAGCAACUCUGAAGCUGUCAACUGACAGUUAUGAGAAAAAGUAAAGCUGACGAAGACAUCAGCGGGGCAAAUUUGUUUAGUCUUAGUCUCUGUCU-CAGUCUUCGUUU--U-CC- --.........(((...(((((((....)))))))..)))..........(..(((((((((....(((((((.................))).))))..-..))))))))).--.-).- ( -28.63) >consensus __AA_AAGCAACUCUGAAGCUGUCAACUGACAGUUAUGAGAAAAAGUAAAGCUGACGAAGACAUCAGCGGGGCAAAUUUGUUUAGUCUUAGUCUCUCUCU_CA__CU_CGUUU__U_CC_ .....(((((((((...(((((((....)))))))..))).....((...(((((........)))))...))....))))))..................................... (-18.01 = -18.65 + 0.64)


| Location | 7,603,240 – 7,603,346 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.77 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7603240 106 + 22224390 CAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUU-UU-----CCAGCCAGCCAGAGUUUUUCU---CCUCUGUC----- .......(((..(((((((....)))))))...................(((....))))))..(((((..(((.-..-----..)))......(((.....))---))))))..----- ( -25.30) >DroPse_CAF1 27151 112 + 1 CAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCUUUUU-----CCAUCCAGCCAGAGCUGUCCU---CCGUUUCCGCCUC .......((...(((((((....)))))))..........................((((((((..(.((((......-----.....))))).))))))))..---........))... ( -23.20) >DroSim_CAF1 21080 100 + 1 CAAAUUUGCCCCGGGGAUGUGUUUGUCAG-UUUAUUUUUUUUCAUAACUGUCAGUUGACGGUUUCAGAGUCGCUU-UU-----CCAGCCAGCCAGAGUUUUUCU---CCU---------- ............(((((....((((.(..-..........(((..(((((((....)))))))...)))..(((.-..-----..)))..).)))).....)))---)).---------- ( -17.60) >DroWil_CAF1 27511 114 + 1 CAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCCU-UUUUUUUCUCGCUCACUCUCGUCCUUGU---CCUCAAU--GUUG ............(((((((....))))))).................(((((....)))))....(((((.((..-..........))..))))).........---.......--.... ( -19.90) >DroYak_CAF1 21640 109 + 1 CAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUU-UUCCUUUCCCGCCAGCCAGAGUUUUUCCACUCCU---------- .......(((..(((((((....)))))))...................(((....))))))....(((..((((-(..((........))..)))))..))).......---------- ( -20.60) >DroPer_CAF1 27031 112 + 1 CAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCUUUUU-----CCAUCCAGCCAGAGUUGUCCU---CCGUUUCCGCCUC .......((...(((((((....)))))))..........................((((((((..(.((((......-----.....))))).))))))))..---........))... ( -20.80) >consensus CAAAUUUGCCCCGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUCGCUU_UU_____CCAGCCAGCCAGAGUUUUUCU___CCU_U________ .......((...(((((((....)))))))..........(((....(((((....))))).....)))..))............................................... (-15.79 = -15.77 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:01 2006