
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,600,357 – 7,600,489 |
| Length | 132 |
| Max. P | 0.723179 |

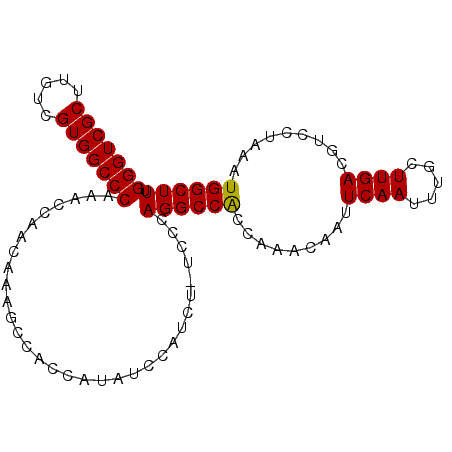
| Location | 7,600,357 – 7,600,450 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -18.98 |
| Energy contribution | -18.79 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7600357 93 - 22224390 GGGUCGCUUGUCGUGGCCCAAACCAACAAUGCCACCAUAUCCAUCU-UCCCAGGCCACCA-ACAAUUCAAUUUGCUUGACGUCCUAAAUGGCUUU (((((((.....)))))))...........................-....((((((...-.....((((.....)))).........)))))). ( -18.53) >DroSec_CAF1 17403 94 - 1 GGGUCGCUUGUCGUGGCCCAAACCAACAAAGCCACCAUAUCCAUCU-UCCCAGGCCACCAAACAAUUCAAUUUGCUUGACGUCCUAAAUGGCUUU (((((((.....)))))))........(((((((............-....(((.(..(((.(((......))).)))..).)))...))))))) ( -21.41) >DroSim_CAF1 17987 94 - 1 GGGUCGCUUGUCGUGGCCCAAACCAACAAAGCCACCAUAUCCAUCU-UCCCAGGCCACCAAACAAUUCAAUUUGCUUGACGUCCUAAAUGGCUUU (((((((.....)))))))........(((((((............-....(((.(..(((.(((......))).)))..).)))...))))))) ( -21.41) >DroEre_CAF1 23097 93 - 1 GGGUCGCUUGUCGUGGCCCAAAUCACCA-AGCAAGCAUAUCCAUUUUUCCCAGGCCGCC-AACAAUUCAAUUUGCUUGACGUCCUAAAUGGCUUU (((((((.....))))))).........-...........((((((.....(((.((.(-(((((......))).))).)).))))))))).... ( -22.90) >consensus GGGUCGCUUGUCGUGGCCCAAACCAACAAAGCCACCAUAUCCAUCU_UCCCAGGCCACCAAACAAUUCAAUUUGCUUGACGUCCUAAAUGGCUUU (((((((.....)))))))................................((((((.........((((.....)))).........)))))). (-18.98 = -18.79 + -0.19)


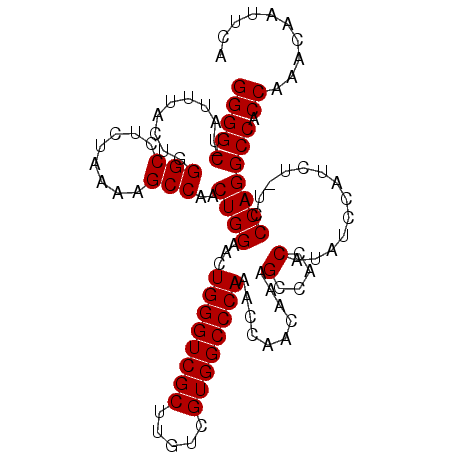
| Location | 7,600,383 – 7,600,489 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -23.65 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7600383 106 - 22224390 GGGGACUAUUUACUGGGCCUCUAAAAGCCAACUGGAACUGGGUCGCUUGUCGUGGCCCAAACCAACAAUGCCACCAUAUCCAUCU-UCCCAGGCCACCA-ACAAUUCA (((((..........(((........)))...((((..((((((((.....))))))))........(((....))).))))..)-)))).........-........ ( -26.00) >DroSec_CAF1 17429 106 - 1 GGGG-CUAUUUACUGGGCCUCUAAAAGCCAACUGGAACUGGGUCGCUUGUCGUGGCCCAAACCAACAAAGCCACCAUAUCCAUCU-UCCCAGGCCACCAAACAAUUCA ((((-((........(((........)))...((((....(((.((((....(((......)))...)))).)))...))))...-.....)))).)).......... ( -28.70) >DroSim_CAF1 18013 106 - 1 GGGG-CUAUUUACUGGGCCUCUAAAAGCCAACUGGAACUGGGUCGCUUGUCGUGGCCCAAACCAACAAAGCCACCAUAUCCAUCU-UCCCAGGCCACCAAACAAUUCA ((((-((........(((........)))...((((....(((.((((....(((......)))...)))).)))...))))...-.....)))).)).......... ( -28.70) >DroEre_CAF1 23123 105 - 1 GGGG-CUAUUUACUGGGCCUCUAAAAGCCAACUGGAACUGGGUCGCUUGUCGUGGCCCAAAUCACCA-AGCAAGCAUAUCCAUUUUUCCCAGGCCGCC-AACAAUUCA ((((-(.........(((........)))..((((((.(((((.((((((.((((......))))..-.))))))..))))).))...))))))).))-......... ( -29.80) >consensus GGGG_CUAUUUACUGGGCCUCUAAAAGCCAACUGGAACUGGGUCGCUUGUCGUGGCCCAAACCAACAAAGCCACCAUAUCCAUCU_UCCCAGGCCACCAAACAAUUCA ((((.(.........(((........)))..((((...((((((((.....))))))))..........(....).............))))))).)).......... (-23.65 = -23.65 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:58 2006