
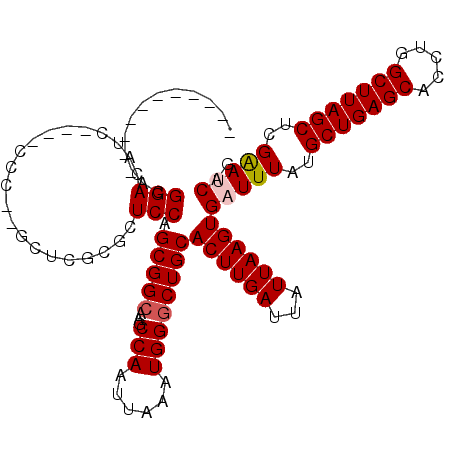
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,597,954 – 7,598,070 |
| Length | 116 |
| Max. P | 0.732532 |

| Location | 7,597,954 – 7,598,053 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7597954 99 + 22224390 -----------ACGGACAUC----CCC--GCUUUCGCUCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAG -----------.(((.....----.))--)....(((....)))(((.(((.........))))))((((((...))))))(((((..(((((((.....)))))))..))))).. ( -30.30) >DroPse_CAF1 17991 96 + 1 -----------GCGGACAUC---------GCCCGGGGUCCAGCGGCAAGCCAAUUAAAUGGACUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCAAGGAACCA -----------.(((.....---------..)))((.(((....(((..(((......)))..)))((((((...))))))......((((((((.....)))))))).))).)). ( -29.80) >DroSim_CAF1 15783 99 + 1 -----------ACGGACAUC----CCC--GCUUUCGCUCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAG -----------.(((.....----.))--)....(((....)))(((.(((.........))))))((((((...))))))(((((..(((((((.....)))))))..))))).. ( -30.30) >DroYak_CAF1 16563 109 + 1 GCAACGGACA-ACGGACAGC----CCC--GCUUUUGCUCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUGGAAUCAG ........((-(.(..((((----((.--(((........)))((....))........))))))..)))).........((((((..(((((((.....)))))))..)))))). ( -33.70) >DroAna_CAF1 15671 111 + 1 -----GGACACAGGGACACCGGGACCGGGACACGAGGUCCAGCGGCAGGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGGAUCCG -----.......((....)).((((((((...(.((((((((((((((.(((......))).))))((((((...))))))......))))).).)))).).....))))).))). ( -40.10) >DroPer_CAF1 18540 96 + 1 -----------GCGGACAUC---------GCCCGGGGUCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCAAGGAACCA -----------.(((.....---------..)))((.(((.(((((...(((......))))))))((((((...))))))......((((((((.....)))))))).))).)). ( -32.20) >consensus ___________ACGGACAUC____CCC__GCUCGCGCUCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAG .............(((.....................))).(((((...(((......))))))))((((((...))))))(((((..(((((((.....)))))))..))))).. (-24.50 = -24.75 + 0.25)


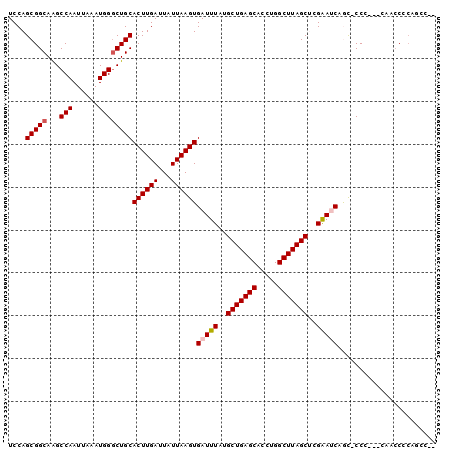
| Location | 7,597,974 – 7,598,070 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7597974 96 + 22224390 UCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAGCACCC---CCACCACAGCC-C ....((((....))........(((.(((((((((...))))))(((((..(((((((.....)))))))..))))).))).))---).......)).-. ( -30.00) >DroPse_CAF1 18008 94 + 1 UCCAGCGGCAAGCCAAUUAAAUGGACUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCAAGGAACCAC-CCG---CAUCCCCAGCC-- ....((((((..(((......)))..))).............(((.(((.((((((((.....))))))))..))).)))-.))---)..........-- ( -26.10) >DroSec_CAF1 15236 96 + 1 UCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAGU-CCC---CCACCUCACCCCU ....(((((...(((......))))))))((((((...))))))(((((..(((((((.....)))))))..)))))...-...---............. ( -26.40) >DroSim_CAF1 15803 96 + 1 UCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAGU-CCC---CAACCUCACCGCC ....((((..((..........(((((((((((((...)))))))((((..(((((((.....)))))))..))))))))-)).---....))..)))). ( -28.66) >DroAna_CAF1 15703 93 + 1 UCCAGCGGCAGGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGGAUCCGC-CCCUCCGG--C----GCCG .......((((.(((......))).))))((((((...))))))(((((..(((((((.....)))))))..)))))(((-(.....))--)----)... ( -32.60) >DroPer_CAF1 18557 94 + 1 UCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCAAGGAACCAC-CCA---CAUCCCCAGCC-- ....((((....)).......((((...(((((((...))))))).....((((((((.....)))))))).((......-)).---....)))))).-- ( -26.60) >consensus UCCAGCGGCAAGCCAAUUAAAUGGGCUGCACUUGAUUAUUAAGUGAUUUAUGCUGAGCACCUGGCUUAGCUCGAAUCAGC_CCC___CAACCCCAGCC__ ....(((((...(((......))))))))((((((...))))))(((((..(((((((.....)))))))..)))))....................... (-23.42 = -23.67 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:57 2006