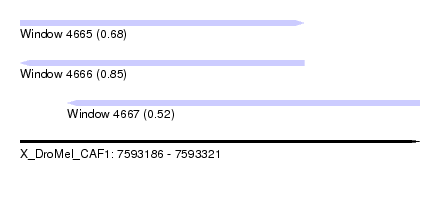
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,593,186 – 7,593,321 |
| Length | 135 |
| Max. P | 0.852688 |

| Location | 7,593,186 – 7,593,282 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7593186 96 + 22224390 -------UUG---UGUGCAUUUAAUGCGAUCCACUGAUAAGCCGACCGGCAAUCGUCAUCGCAAUCGGAAUGCAAUUGCAAUUGCCAUUGUCAGCAAUUCAAUUGC -------...---..(((((((..((((((....((((..(((....))).))))..))))))....)))))))...(((((((..((((....)))).))))))) ( -28.10) >DroSim_CAF1 11046 96 + 1 -------UUG---UGUGCAUUUAAUGCGAUCCGCUGAUAAGCCGACCGGCAAUCGUCAUCGCAAUCGGAAUGCAAUUGCAAUUGCCAUUGUCAGCAAUUCAAUUGC -------...---..(((((((..((((((..((.(((..(((....))).))))).))))))....)))))))...(((((((..((((....)))).))))))) ( -28.20) >DroEre_CAF1 12139 96 + 1 -------UUG---UGUGCAUUUAAUGCGAUCCGCUGAUAAGCCGACCGGCAAUCGUCAUCGCAAUCGAAAUGCAAUUGCAAUUGCCAUUGUCAGCAAUUCAAUUGC -------...---..(((((((..((((((..((.(((..(((....))).))))).))))))....)))))))...(((((((..((((....)))).))))))) ( -27.80) >DroYak_CAF1 10781 96 + 1 -------UUG---UGUGCAUUUAAUGCGACCCGCUGAUAAGCCGACCGGCAAUCGUCAUCGCAAUCGGAAUGCAAUUGCAAUUGCCAUUGUCAGCAAUUCAAUUGC -------...---..(((((((..(((((...((.(((..(((....))).)))))..)))))....)))))))...(((((((..((((....)))).))))))) ( -27.40) >DroAna_CAF1 9963 94 + 1 UCCUUUCUUGGCUGGAGCGUUUAAUGCGAACCGCUGAUAAGCCGCCCGGCAAU------CGCAAUUGGAAUGCAAUUGCAAU------UGUCAGCAAUUCAAUUGC .((......))..((..(((.....)))..))(((((((((((....)))...------.(((((((.....)))))))..)------)))))))........... ( -30.00) >DroPer_CAF1 12242 94 + 1 UUGU---UUC---UCUGCGUUUAAUGUGAGCCACUGAUAAGCCGCGCGGCAAUCGAAAACGCAAUUGAAAUGCAAUUGCA------AUUGUCAGCAAUUCAAUUGA ..((---(((---..(((((((...(((...)))((((..(((....))).)))).)))))))...))))).((((((.(------((((....))))))))))). ( -25.10) >consensus _______UUG___UGUGCAUUUAAUGCGAUCCGCUGAUAAGCCGACCGGCAAUCGUCAUCGCAAUCGGAAUGCAAUUGCAAUUGCCAUUGUCAGCAAUUCAAUUGC ................((((...))))((...(((((((((((....)))..........(((((.(.....).)))))........))))))))...))...... (-20.19 = -20.33 + 0.14)



| Location | 7,593,186 – 7,593,282 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -22.69 |
| Energy contribution | -23.73 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7593186 96 - 22224390 GCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGUGGAUCGCAUUAAAUGCACA---CAA------- ...(((((...((((((...((((((((..((((((.......))))))..)))))))).))))))...))))).....((((...))))...---...------- ( -34.50) >DroSim_CAF1 11046 96 - 1 GCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGAUCGCAUUAAAUGCACA---CAA------- (((..(((...((((((...((((((((..((((((.......))))))..)))))))).))))))...)))(((...)))......)))...---...------- ( -35.30) >DroEre_CAF1 12139 96 - 1 GCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUUCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGAUCGCAUUAAAUGCACA---CAA------- (((..(((...((((((...((((((((..((((((.......))))))..)))))))).))))))...)))(((...)))......)))...---...------- ( -35.30) >DroYak_CAF1 10781 96 - 1 GCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGGUCGCAUUAAAUGCACA---CAA------- (((..(((...((((((...((((((((..((((((.......))))))..)))))))).))))))...)))(((...)))......)))...---...------- ( -35.30) >DroAna_CAF1 9963 94 - 1 GCAAUUGAAUUGCUGACA------AUUGCAAUUGCAUUCCAAUUGCG------AUUGCCGGGCGGCUUAUCAGCGGUUCGCAUUAAACGCUCCAGCCAAGAAAGGA ((....((((((((((.(------((((((((((.....))))))))------)))(((....)))...))))))))))((.......))....)).......... ( -31.10) >DroPer_CAF1 12242 94 - 1 UCAAUUGAAUUGCUGACAAU------UGCAAUUGCAUUUCAAUUGCGUUUUCGAUUGCCGCGCGGCUUAUCAGUGGCUCACAUUAAACGCAGA---GAA---ACAA .(((((((((((....))))------).))))))..((((..((((((((......(((((...........))))).......)))))))).---)))---)... ( -24.12) >consensus GCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGAUCGCAUUAAAUGCACA___CAA_______ (((.((((..(((.(.((((.(..((((((((((.....))))))))))..).)))).)((((.(((....))).))))))))))).)))................ (-22.69 = -23.73 + 1.04)


| Location | 7,593,202 – 7,593,321 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.60 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -25.81 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7593202 119 - 22224390 -AUUGUGGACUUCUUAGAAGACAAACGGCUCAAUAAAGUCGCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGUGGAUCG -.((((...((((...)))))))).(((((......)))))..(((((...((((((...((((((((..((((((.......))))))..)))))))).))))))...)))))...... ( -38.10) >DroSec_CAF1 10494 119 - 1 -AUUGUGGACUUCUUCGAAGACAAACGGCUCAAUAAAGUCGCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGAUCG -.((((...((((...))))))))..(((........(((((((((((((((((......)))))))((....))....)))))))))).......)))((((.(((....))).)))). ( -37.76) >DroEre_CAF1 12155 119 - 1 -AUUGUGGACUUAUCGGAAGACAAACGGCUCAAUAAAGUCGCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUUCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGAUCG -............((....)).....(((........((((((((((((.(((((.((((....))))))...))).)))))))))))).......)))((((.(((....))).)))). ( -39.26) >DroYak_CAF1 10797 119 - 1 -AUUGUGGACUUCUUGGAAGACAAACGGCUCAAUAAAGUCGCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGGUCG -......((((..((((....(((.(((((......)))))...)))....((((((...((((((((..((((((.......))))))..)))))))).))))))...))))..)))). ( -40.70) >DroAna_CAF1 9989 108 - 1 GAAUUCUGAGUGGGCACAAGACAAACGGCCCAAUAAAGUCGCAAUUGAAUUGCUGACA------AUUGCAAUUGCAUUCCAAUUGCG------AUUGCCGGGCGGCUUAUCAGCGGUUCG (((((((((.(((((............)))))...(((((((..........(((.((------((((((((((.....))))))))------)))).)))))))))).)))).))))). ( -38.40) >DroPer_CAF1 12262 108 - 1 -AUGGUAGA-----UGGUAGACAGACGACUCAAUAAAGUCUCAAUUGAAUUGCUGACAAU------UGCAAUUGCAUUUCAAUUGCGUUUUCGAUUGCCGCGCGGCUUAUCAGUGGCUCA -..(((((.-----(((.((((....((((......)))).((((((((.(((.......------.......))).)))))))).))))))).)))))(.((.(((....))).)).). ( -28.14) >consensus _AUUGUGGACUUCUUGGAAGACAAACGGCUCAAUAAAGUCGCAAUUGAAUUGCUGACAAUGGCAAUUGCAAUUGCAUUCCGAUUGCGAUGACGAUUGCCGGUCGGCUUAUCAGCGGAUCG ..........................(((........((((((((((...(((((.((((....))))))...)))...)))))))))).......)))((((.(((....))).)))). (-25.81 = -26.48 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:54 2006