
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,579,910 – 7,580,005 |
| Length | 95 |
| Max. P | 0.651738 |

| Location | 7,579,910 – 7,580,005 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -25.36 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7579910 95 + 22224390 ------UUGGGCGGCGGUGCCAGAC----------------UGGCGGUGUUGCCAUUAGCCGGUUACCGUUUUCAAGCCAAGUCUAGCUAGUUUUUGUGUACCGCCGAAAAAUACAU ------.....((((((((((((((----------------((((((((..(((.......)))))))(((....)))........)))))))..)).))))))))).......... ( -34.00) >DroSec_CAF1 2703 111 + 1 ------UUGGGCGGCGGUGCCAGACUGGCGGCGGGCCAGAUUAGUGGUGUUGCCAUUAGCCGGUUACCGUUUUCAAGCCAAAUCUAGCUAGCUUUUGUGUACCGCCGAAAUAUACCU ------..((.(((((((((((((..(((((((((((.(.(((((((.....)))))))).)))..)))))....(((........))).))))))).))))))))).......)). ( -41.10) >DroSim_CAF1 2892 95 + 1 ------UUGGGCGGCGGUGCCAGAC----------------UAGUGGUGUUGCCAUUAGCCGGCUACCGUUUUCAAGCCAAAUCUAGCUAGCUUUUGUGUACCGCCGAAAUAUACCU ------..((.((((((((((((((----------------((((((.....)))))))..((((..........))))..............)))).))))))))).......)). ( -34.10) >DroEre_CAF1 2715 101 + 1 UGACAUGUGGGUGGCGGUGCCAGAC----------------UCGCUGCAUUGCCUCUAACCGGUUAUCGUUUUCAAGCCAAAUCUAGCUGGAUUUUGUGUAUAGCUAAAAAAUACAA ..((((.((((.((((((((.((..----------------...)))))))))))))).(((((((..((((.......)))).))))))).....))))................. ( -23.90) >consensus ______UUGGGCGGCGGUGCCAGAC________________UAGCGGUGUUGCCAUUAGCCGGUUACCGUUUUCAAGCCAAAUCUAGCUAGCUUUUGUGUACCGCCGAAAAAUACAU ...........(((((((((((((..................(((((((..(((.......))))))))))....(((........)))....)))).))))))))).......... (-25.36 = -24.68 + -0.69)



| Location | 7,579,910 – 7,580,005 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.15 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7579910 95 - 22224390 AUGUAUUUUUCGGCGGUACACAAAAACUAGCUAGACUUGGCUUGAAAACGGUAACCGGCUAAUGGCAACACCGCCA----------------GUCUGGCACCGCCGCCCAA------ ..........(((((((............((((((((.(((.......(((...))).....((....))..))))----------------)))))))))))))).....------ ( -32.60) >DroSec_CAF1 2703 111 - 1 AGGUAUAUUUCGGCGGUACACAAAAGCUAGCUAGAUUUGGCUUGAAAACGGUAACCGGCUAAUGGCAACACCACUAAUCUGGCCCGCCGCCAGUCUGGCACCGCCGCCCAA------ ..........(((((((........((..(((((((((((........(((...))).....((....)))))..))))))))..)).(((.....)))))))))).....------ ( -35.90) >DroSim_CAF1 2892 95 - 1 AGGUAUAUUUCGGCGGUACACAAAAGCUAGCUAGAUUUGGCUUGAAAACGGUAGCCGGCUAAUGGCAACACCACUA----------------GUCUGGCACCGCCGCCCAA------ ...........((((((......(((((((......)))))))......(((.((((((((.(((.....))).))----------------).))))))))))))))...------ ( -35.50) >DroEre_CAF1 2715 101 - 1 UUGUAUUUUUUAGCUAUACACAAAAUCCAGCUAGAUUUGGCUUGAAAACGAUAACCGGUUAGAGGCAAUGCAGCGA----------------GUCUGGCACCGCCACCCACAUGUCA .(((((.........))))).........(((((((((((((((....(..(((....)))..).))).))..)))----------------))))))).................. ( -18.00) >consensus AGGUAUAUUUCGGCGGUACACAAAAGCUAGCUAGAUUUGGCUUGAAAACGGUAACCGGCUAAUGGCAACACCACUA________________GUCUGGCACCGCCGCCCAA______ ..........(((((((............(((((((((((((.(..........).))))))((....))......................))))))))))))))........... (-19.27 = -19.15 + -0.12)

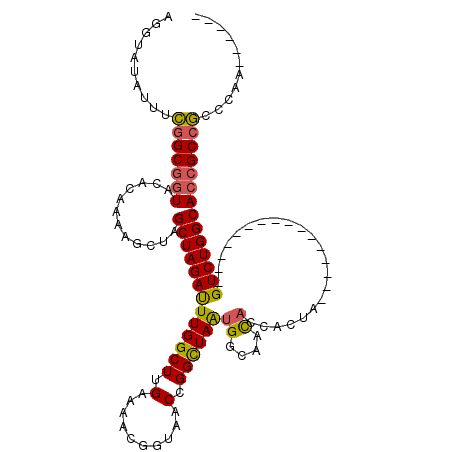

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:52 2006