| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,571,923 – 7,572,021 |
| Length | 98 |
| Max. P | 0.608470 |

| Location | 7,571,923 – 7,572,021 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.76 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -9.42 |
| Energy contribution | -8.90 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
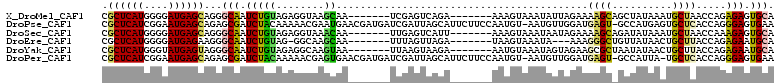
>X_DroMel_CAF1 7571923 98 + 22224390 CGCUCAUGGGGAUGAGCAGGGCAAUCUGUAGAGGUAAGCAA-------UCGAGUCAGA-------AAAGUAAAUAUUAGAAAAGCAGCUAUAAAUGCUAACCAGAGAGUGCA .((((((....))))))...(((.(((.(...(((.((((.-------((......))-------...........(((........)))....)))).)))).))).))). ( -21.50) >DroPse_CAF1 2406 110 + 1 CGCUCAUCGGAAUGAGCAGAGCGAUCUACAAAAACGAAUGAACGAUGAUCGAUUAGCAUUCUUCCAAUGU-AAUGUUGGAUGAGU-GCCAUGAGUGCUCACCAGGGAGUGAA (((((...((..((((((...(((((....................)))))....((((((.((((((..-...)))))).))))-))......))))))))...))))).. ( -35.95) >DroSec_CAF1 1840 98 + 1 CGCUCAUGGGGAUGAGCAGGGCAAUCUGUAGAGGUAAACAA-------UUGAGUCAUU-------AAAGUAAAUAAUAGAAAAGCAGAUAUAAAUGCUAACCAAAGAGUGCA .((((((....))))))...(((.(((.....(((......-------((((....))-------))...............((((........)))).)))..))).))). ( -19.20) >DroEre_CAF1 11400 94 + 1 CGCUCAUGGGGAUGAGAAGGGCAAUCUGUAG-GGCAAGCAA-------UUUAGUUAGA-------UAAGUAAAUA---AAAGGGCUGUUAUAACUGCUUACCAGAGAAUGCA ..(((((....)))))....(((.(((.(.(-(..(((((.-------....((((..-------..(((.....---.....)))....))))))))).))).))).))). ( -20.30) >DroYak_CAF1 10008 98 + 1 CGCUCAUGGGUAUGAGUAGGGCAAUCUGUAGAGGCAAGUAA-------UUAAGUAAGA-------AAUGUAAAUAGUAGAAGCGCUAAUAUAACUGCUUACCAGAGAAUGCA .((((((....))))))...(((.((((...(((((.((..-------.......(..-------..).....((((......)))).....)))))))..))))...))). ( -18.20) >DroPer_CAF1 2385 109 + 1 CGCUCAUCGGAAUGAGCAGAGCGAUCUACAAAAACGAGUGAACGAUGAUCGAUUAGCAUUCUUCCAAUGU-AAUGUUGGAUGAGU-GCCAUUA-UGCUCACCAGGGAGUGAA (((((...((..((((((...(((((....................)))))....((((((.((((((..-...)))))).))))-)).....-))))))))...))))).. ( -36.05) >consensus CGCUCAUGGGGAUGAGCAGGGCAAUCUGUAGAGGCAAGCAA_______UCGAGUAAGA_______AAAGUAAAUAUUAGAAGAGC_GCUAUAAAUGCUAACCAGAGAGUGCA .((((((....))))))...(((.(((((........))..........................................((((..........)))).....))).))). ( -9.42 = -8.90 + -0.52)

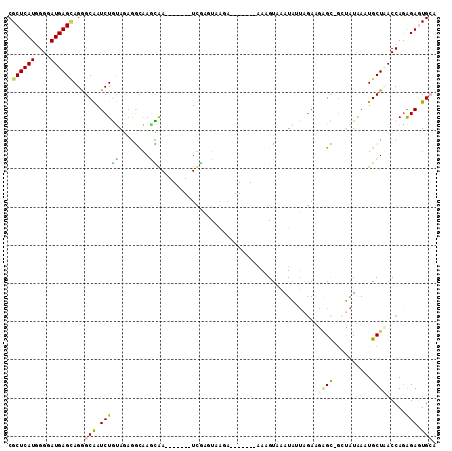
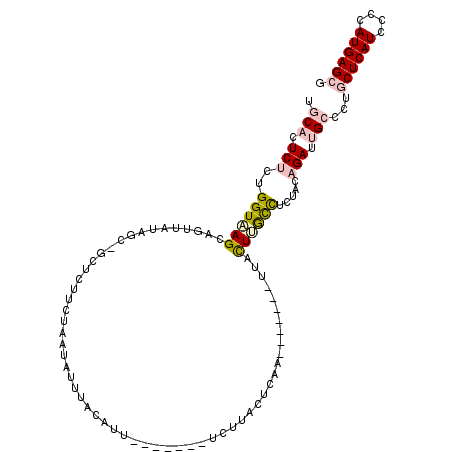
| Location | 7,571,923 – 7,572,021 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.76 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -7.26 |
| Energy contribution | -7.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7571923 98 - 22224390 UGCACUCUCUGGUUAGCAUUUAUAGCUGCUUUUCUAAUAUUUACUUU-------UCUGACUCGA-------UUGCUUACCUCUACAGAUUGCCCUGCUCAUCCCCAUGAGCG .(((.(((..(((.((((....(((........)))...........-------((......))-------.)))).))).....))).)))...((((((....)))))). ( -17.30) >DroPse_CAF1 2406 110 - 1 UUCACUCCCUGGUGAGCACUCAUGGC-ACUCAUCCAACAUU-ACAUUGGAAGAAUGCUAAUCGAUCAUCGUUCAUUCGUUUUUGUAGAUCGCUCUGCUCAUUCCGAUGAGCG ..........(((((.......((((-(.((.(((((....-...))))).)).))))).....)))))(((((((.(.....(((((....))))).....).))))))). ( -28.70) >DroSec_CAF1 1840 98 - 1 UGCACUCUUUGGUUAGCAUUUAUAUCUGCUUUUCUAUUAUUUACUUU-------AAUGACUCAA-------UUGUUUACCUCUACAGAUUGCCCUGCUCAUCCCCAUGAGCG .(((.(((.(((..((((........))))......(((((......-------))))).....-------..........))).))).)))...((((((....)))))). ( -16.00) >DroEre_CAF1 11400 94 - 1 UGCAUUCUCUGGUAAGCAGUUAUAACAGCCCUUU---UAUUUACUUA-------UCUAACUAAA-------UUGCUUGCC-CUACAGAUUGCCCUUCUCAUCCCCAUGAGCG .(((.(((..(((((((((((.............---..........-------........))-------)))))))))-....))).)))....(((((....))))).. ( -19.58) >DroYak_CAF1 10008 98 - 1 UGCAUUCUCUGGUAAGCAGUUAUAUUAGCGCUUCUACUAUUUACAUU-------UCUUACUUAA-------UUACUUGCCUCUACAGAUUGCCCUACUCAUACCCAUGAGCG .(((.(((..((((((.(((((.........................-------.......)))-------)).)))))).....))).)))....(((((....))))).. ( -15.45) >DroPer_CAF1 2385 109 - 1 UUCACUCCCUGGUGAGCA-UAAUGGC-ACUCAUCCAACAUU-ACAUUGGAAGAAUGCUAAUCGAUCAUCGUUCACUCGUUUUUGUAGAUCGCUCUGCUCAUUCCGAUGAGCG ..........(((((...-...((((-(.((.(((((....-...))))).)).))))).....)))))(((((.(((.....(((((....)))))......)))))))). ( -28.80) >consensus UGCACUCUCUGGUAAGCAGUUAUAGC_GCUCUUCUAAUAUUUACAUU_______UCUUACUCAA_______UUACUUGCCUCUACAGAUUGCCCUGCUCAUCCCCAUGAGCG .(((.(((..((((((..........................................................)))))).....))).)))...((((((....)))))). ( -7.26 = -7.82 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:49 2006