| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,567,276 – 7,567,390 |
| Length | 114 |
| Max. P | 0.961858 |

| Location | 7,567,276 – 7,567,390 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -22.35 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
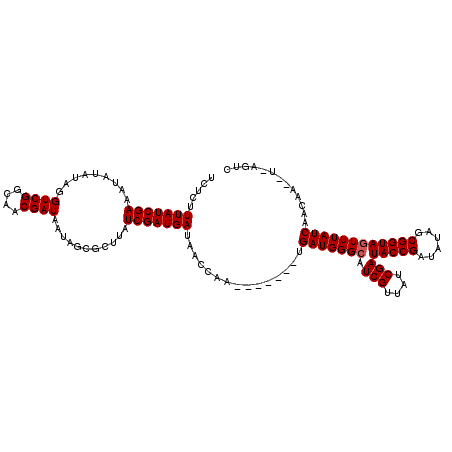
>X_DroMel_CAF1 7567276 114 + 22224390 UCUCUUUAUCGAAA------AGUCGGCAACGAUAAUAGCGCCUAUCGAUGACGAUCAAGAAUAGGUGAUGGGCAUCGUUACCGAUACCGAUAUAGCGGUAGUUUAUCAACAAUCUCAGUC .....(((((((..------.((((....))))..(((...)))))))))).(((..(((.....((((((((.(((....)))(((((......)))))))))))))....)))..))) ( -28.10) >DroSec_CAF1 59681 113 + 1 UCUCUUUAUCGAAAUAUAUAGGUCGGCAACGAUAAUAGCGCUUAUCGAUGAUAAGCGA-------UGAUGGGCAUCGUUAUCGAUACCGAUAUAGCGGUAGUUUAUCAACAACAUUAGUC .....................((((....)))).....((((((((...)))))))).-------((((((((((((((((((....)))))..))))).))))))))............ ( -31.40) >DroSim_CAF1 53760 113 + 1 UCUCUUUAUCGAAAUAUAUAGGUCGGCCACGAUAAUAGUGCUUAUCGAUGAUAACCAA-------GGAUGGGCAUCGUUAUCGAUACCGAAAAACCGGUAAUUUAUCAACANNNNNNNNN ....................((....)).((((((..(((((((((..((.....)).-------.)))))))))..)))))).(((((......))))).................... ( -27.70) >consensus UCUCUUUAUCGAAAUAUAUAGGUCGGCAACGAUAAUAGCGCUUAUCGAUGAUAACCAA_______UGAUGGGCAUCGUUAUCGAUACCGAUAUAGCGGUAGUUUAUCAACAA__U_AGUC .....(((((((.........((((....))))...........)))))))...............(((((((.(((....)))(((((......))))))))))))............. (-22.35 = -22.68 + 0.33)


| Location | 7,567,276 – 7,567,390 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
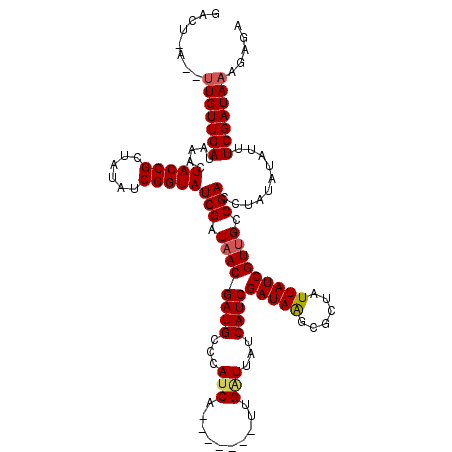
>X_DroMel_CAF1 7567276 114 - 22224390 GACUGAGAUUGUUGAUAAACUACCGCUAUAUCGGUAUCGGUAACGAUGCCCAUCACCUAUUCUUGAUCGUCAUCGAUAGGCGCUAUUAUCGUUGCCGACU------UUUCGAUAAAGAGA ........(((((((.....(((((......)))))((((((((((((.....(.((((((..((.....))..)))))).)....))))))))))))..------..)))))))..... ( -33.90) >DroSec_CAF1 59681 113 - 1 GACUAAUGUUGUUGAUAAACUACCGCUAUAUCGGUAUCGAUAACGAUGCCCAUCA-------UCGCUUAUCAUCGAUAAGCGCUAUUAUCGUUGCCGACCUAUAUAUUUCGAUAAAGAGA ..((....(((((((...............((((((.((((((.((((.....))-------))(((((((...)))))))....)))))).))))))..........)))))))..)). ( -27.61) >DroSim_CAF1 53760 113 - 1 NNNNNNNNNUGUUGAUAAAUUACCGGUUUUUCGGUAUCGAUAACGAUGCCCAUCC-------UUGGUUAUCAUCGAUAAGCACUAUUAUCGUGGCCGACCUAUAUAUUUCGAUAAAGAGA .........((((((.....((..((((....(((((((....))))))).....-------..(((((....((((((......)))))))))))))))..))....))))))...... ( -24.50) >consensus GACU_A__UUGUUGAUAAACUACCGCUAUAUCGGUAUCGAUAACGAUGCCCAUCA_______UUGAUUAUCAUCGAUAAGCGCUAUUAUCGUUGCCGACCUAUAUAUUUCGAUAAAGAGA ........(((((((.....(((((......)))))(((.((((((((...(((..........)))...))))(((((......))))))))).)))..........)))))))..... (-18.10 = -18.77 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:45 2006