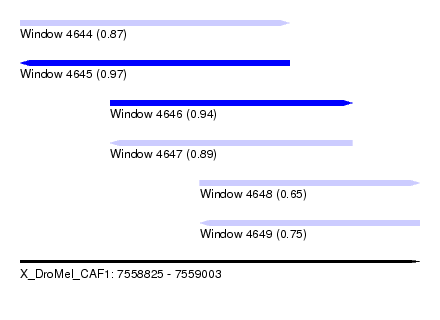
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,558,825 – 7,559,003 |
| Length | 178 |
| Max. P | 0.965006 |

| Location | 7,558,825 – 7,558,945 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -56.02 |
| Consensus MFE | -43.06 |
| Energy contribution | -45.34 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7558825 120 + 22224390 GUAGGGUGCAUAAAGUGGUGGAUACCAAACCGGUGGUGGUGGCGGCAGGACCAAAUCCUGUGGUGGCAACGAUGCCGCCGGCGGUGUAUUGUCCUGCAGGUUAGUGUCGACGGCACCACC ....(((((......((((....))))...((.((..(.((((.(((((((...(..(((((((((((....))))))).))))..)...)))))))..)))).)..)).)))))))... ( -50.20) >DroPse_CAF1 115708 120 + 1 GUACGGGGCAUACAGCGGGGGAUACCACACGGGUGGUGGCGGUGGCAGGACCAAAUCCUGUGGCGGUAACGAUGCCGCCGGCGGUGUGUUGUCCUGCAUUGAACUGUCCACCGCUCCGCC ...((((((..((..((.((....))...)).))(((((((((.(((((((...(..(((((((((((....))))))).))))..)...))))))).....)))).))))))))))).. ( -58.80) >DroEre_CAF1 62811 120 + 1 GUAGGGCGCGUAGAGCGGUGGAUACCAAACCGGUGGUGGUGGCGGCAGGACCAAAUCCUGUGGUGGCAACGAUGCCGCUGGCGGCGUAUUAUCCUGCAGGUUAGUGUCCACCGCACCACC ...((((((.....))(((((((((..((((.(..(.(((.((.((((((.....)))))).)).))....((((((....)))))).....))..).)))).))))))))))).))... ( -55.50) >DroYak_CAF1 52809 120 + 1 GUAUGGCGCGUACAGCGGUGGAUACCACACCGGUGGCGGUGGCGGCAGCACCAGAUCCUGUGGCGGCAACGAUGCCGCCGGCGGUGCAUUAUCCUGCAGGCUACUGUCCACUGCACCGCC ....((((.((.((.(((((.......))))).))((((((((((.(((........(((((((((((....))))))).))))((((......)))).))).))).))))))))))))) ( -59.00) >DroPer_CAF1 121823 120 + 1 GUACGGGGCAUACAGCGGGGGAUACCACACGGGUGGUGGCGGUGGCAGGACCAAAUCCUGUGGCGGUAAUGAUGCUGCCGGCGGUGUGUUGUCCUGCAUCGAACUGUCCACCGCUCCGCC ...((((((..((..((.((....))...)).))(((((((((.(((((((...(..(((((((((((....))))))).))))..)...))))))).....)))).))))))))))).. ( -56.60) >consensus GUACGGCGCAUACAGCGGUGGAUACCACACCGGUGGUGGUGGCGGCAGGACCAAAUCCUGUGGCGGCAACGAUGCCGCCGGCGGUGUAUUGUCCUGCAGGGUACUGUCCACCGCACCGCC ....(((((.......((......)).....((((((((((((.(((((((...((.(((((((((((....))))))).)))).))...)))))))..))))))).))))))))))... (-43.06 = -45.34 + 2.28)

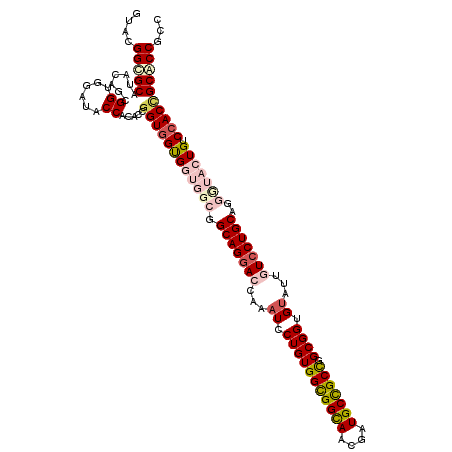
| Location | 7,558,825 – 7,558,945 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -50.26 |
| Consensus MFE | -40.66 |
| Energy contribution | -42.50 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.965006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7558825 120 - 22224390 GGUGGUGCCGUCGACACUAACCUGCAGGACAAUACACCGCCGGCGGCAUCGUUGCCACCACAGGAUUUGGUCCUGCCGCCACCACCACCGGUUUGGUAUCCACCACUUUAUGCACCCUAC ((((((((((.............(((((((......((...((.((((....)))).))...)).....)))))))....(((......))).))))).)))))................ ( -41.10) >DroPse_CAF1 115708 120 - 1 GGCGGAGCGGUGGACAGUUCAAUGCAGGACAACACACCGCCGGCGGCAUCGUUACCGCCACAGGAUUUGGUCCUGCCACCGCCACCACCCGUGUGGUAUCCCCCGCUGUAUGCCCCGUAC (((((.((((((((....))...(((((((......((...(((((........)))))...)).....))))))))))))).(((((....))))).....)))))(((((...))))) ( -49.00) >DroEre_CAF1 62811 120 - 1 GGUGGUGCGGUGGACACUAACCUGCAGGAUAAUACGCCGCCAGCGGCAUCGUUGCCACCACAGGAUUUGGUCCUGCCGCCACCACCACCGGUUUGGUAUCCACCGCUCUACGCGCCCUAC .((((.((((((((.(((((...(((((((......((....(.((((....)))).)....)).....)))))))....(((......)))))))).)))))))).))))......... ( -48.70) >DroYak_CAF1 52809 120 - 1 GGCGGUGCAGUGGACAGUAGCCUGCAGGAUAAUGCACCGCCGGCGGCAUCGUUGCCGCCACAGGAUCUGGUGCUGCCGCCACCGCCACCGGUGUGGUAUCCACCGCUGUACGCGCCAUAC (((((((((((((.(((....)))..(((((..((((((..(((((((....)))))))...((...(((((....)))))...))..))))))..))))).))))))))).)))).... ( -65.10) >DroPer_CAF1 121823 120 - 1 GGCGGAGCGGUGGACAGUUCGAUGCAGGACAACACACCGCCGGCAGCAUCAUUACCGCCACAGGAUUUGGUCCUGCCACCGCCACCACCCGUGUGGUAUCCCCCGCUGUAUGCCCCGUAC (((((.((((((....((((......))))....)))))).(((((.((((...((......))...)))).))))).)))))(((((....))))).......((.....))....... ( -47.40) >consensus GGCGGUGCGGUGGACAGUAACCUGCAGGACAAUACACCGCCGGCGGCAUCGUUGCCGCCACAGGAUUUGGUCCUGCCGCCACCACCACCGGUGUGGUAUCCACCGCUGUAUGCCCCCUAC (((((((((((((..........(((((((......((...(((((((....)))))))...)).....))))))).(((((.((.....))))))).....))))))))).)))).... (-40.66 = -42.50 + 1.84)


| Location | 7,558,865 – 7,558,973 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -48.75 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.95 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7558865 108 + 22224390 GGCGGCAGGACCAAAUCCUGUGGUGGCAACGAUGCCGCCGGCGGUGUAUUGUCCUGCAGGUUAGUGUCGACGGCACCACCCAGACCACCAGUCUUAUUGCCA------GCCG------UU (((.((((((.....))))))(((((((....)))))))((((((((...(((..(((......))).))).)))))....((((.....))))....))).------))).------.. ( -43.80) >DroPse_CAF1 115748 95 + 1 GGUGGCAGGACCAAAUCCUGUGGCGGUAACGAUGCCGCCGGCGGUGUGUUGUCCUGCAUUGAACUGUCCACCGCUCCGCCCAGGCCGCCCUU-------------------G------GU ((((((((((.....))))..(((((((....)))))))(((((.(((.((..(...........)..)).))).)))))...))))))...-------------------.------.. ( -41.60) >DroEre_CAF1 62851 108 + 1 GGCGGCAGGACCAAAUCCUGUGGUGGCAACGAUGCCGCUGGCGGCGUAUUAUCCUGCAGGUUAGUGUCCACCGCACCACCCAGGCCACCAGCCUUGUUGCCA------CCGU------UU ....((((((.....))))))(((((((((((....(((((.((((((......))).(((..((((.....)))).)))...))).))))).)))))))))------))..------.. ( -47.40) >DroYak_CAF1 52849 108 + 1 GGCGGCAGCACCAGAUCCUGUGGCGGCAACGAUGCCGCCGGCGGUGCAUUAUCCUGCAGGCUACUGUCCACUGCACCGCCCAAUGAGGCAGUCUUACUGCCA------GCGC------UU (((((((((..(((...)))..))(....)..)))))))(((((((((.......((((....))))....)))))))))......(((((.....))))).------....------.. ( -50.90) >DroAna_CAF1 54236 120 + 1 GGCGGCAGCACCAGGUCCUGUGGCGGCAGGGAGGCUGCCGGCGGGGUGUUGUCCUGCAGGACGCUGUCCACGGCCCCGCCCAGGGAGGCGUUCUUGCCGCCGUUCGCCGCCGUGGAUGUG (((((((((..(((...))).(((((((((((.(((.((((((((.....((((....))))((((....))))))))))...)).))).)))))))))))))).))))))......... ( -69.00) >DroPer_CAF1 121863 95 + 1 GGUGGCAGGACCAAAUCCUGUGGCGGUAAUGAUGCUGCCGGCGGUGUGUUGUCCUGCAUCGAACUGUCCACCGCUCCGCCCAGGCCGCCUUU-------------------G------GU ((((((((((.....))))..(((((((....)))))))(((((.(((.((..(...........)..)).))).)))))...))))))...-------------------.------.. ( -39.80) >consensus GGCGGCAGGACCAAAUCCUGUGGCGGCAACGAUGCCGCCGGCGGUGUAUUGUCCUGCAGGGUACUGUCCACCGCACCGCCCAGGCCGCCAGUCUU__UGCCA______GC_G______UU (((((((((((......((((((((((......)))))).))))......)))))))........((.....))..))))........................................ (-27.86 = -27.95 + 0.09)

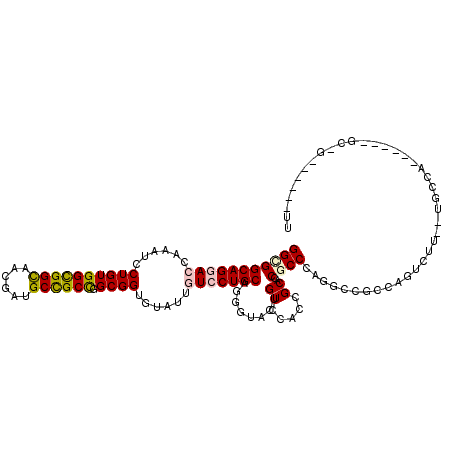
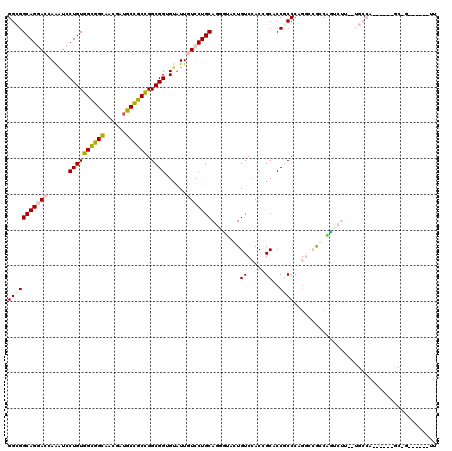
| Location | 7,558,865 – 7,558,973 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -46.54 |
| Consensus MFE | -26.65 |
| Energy contribution | -27.77 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7558865 108 - 22224390 AA------CGGC------UGGCAAUAAGACUGGUGGUCUGGGUGGUGCCGUCGACACUAACCUGCAGGACAAUACACCGCCGGCGGCAUCGUUGCCACCACAGGAUUUGGUCCUGCCGCC ..------.(((------.((((....(((..(...(((((((((((..(((...............)))....)))))))((.((((....)))).)).))))..)..))).))))))) ( -42.26) >DroPse_CAF1 115748 95 - 1 AC------C-------------------AAGGGCGGCCUGGGCGGAGCGGUGGACAGUUCAAUGCAGGACAACACACCGCCGGCGGCAUCGUUACCGCCACAGGAUUUGGUCCUGCCACC ..------.-------------------.(((((..(((((((((.((((((....((((......))))....)))))).((((....)))).))))).)))).....)))))...... ( -39.50) >DroEre_CAF1 62851 108 - 1 AA------ACGG------UGGCAACAAGGCUGGUGGCCUGGGUGGUGCGGUGGACACUAACCUGCAGGAUAAUACGCCGCCAGCGGCAUCGUUGCCACCACAGGAUUUGGUCCUGCCGCC ..------..((------(((((((.((((.....)))).((((.((((((((....((.((....)).)).....))))).))).))))))))))))).((((((...))))))..... ( -49.10) >DroYak_CAF1 52849 108 - 1 AA------GCGC------UGGCAGUAAGACUGCCUCAUUGGGCGGUGCAGUGGACAGUAGCCUGCAGGAUAAUGCACCGCCGGCGGCAUCGUUGCCGCCACAGGAUCUGGUGCUGCCGCC .(------((((------((((((.....)))))((.((((((((((((.((..(.((.....)).)..)).)))))))))(((((((....))))))).)))))...))))))...... ( -53.00) >DroAna_CAF1 54236 120 - 1 CACAUCCACGGCGGCGAACGGCGGCAAGAACGCCUCCCUGGGCGGGGCCGUGGACAGCGUCCUGCAGGACAACACCCCGCCGGCAGCCUCCCUGCCGCCACAGGACCUGGUGCUGCCGCC .........(((((((.((((((.......))))..(((((((((((..((.....))((((....))))....)))))))(((((.....)))))....)))).....))..))))))) ( -58.60) >DroPer_CAF1 121863 95 - 1 AC------C-------------------AAAGGCGGCCUGGGCGGAGCGGUGGACAGUUCGAUGCAGGACAACACACCGCCGGCAGCAUCAUUACCGCCACAGGAUUUGGUCCUGCCACC ..------.-------------------...((((((((((((((.((((((....((((......))))....)))))).((.....))....))))).))))........)))))... ( -36.80) >consensus AA______C_GC______UGGCA__AAGAAUGGCGGCCUGGGCGGUGCGGUGGACAGUAACCUGCAGGACAACACACCGCCGGCGGCAUCGUUGCCGCCACAGGAUUUGGUCCUGCCGCC ....................................(((((((((((...((..(.((.....)).)..))...)))))))(((((((....))))))).))))...(((.....))).. (-26.65 = -27.77 + 1.12)

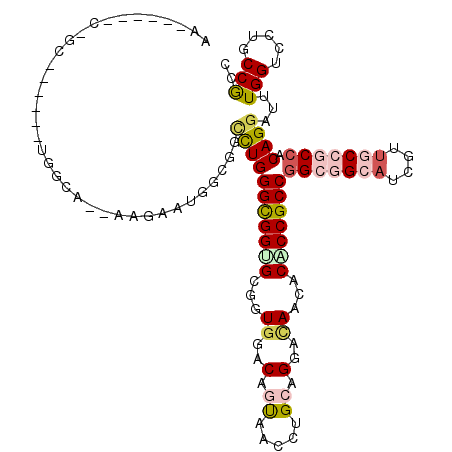
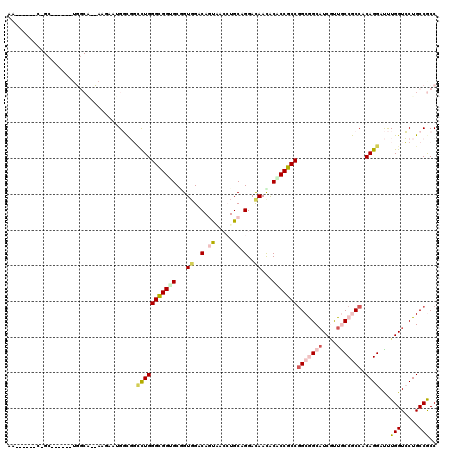
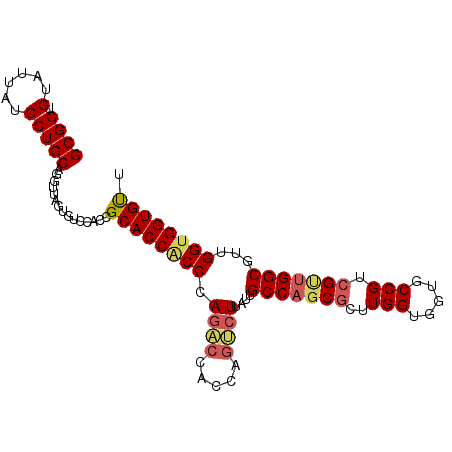
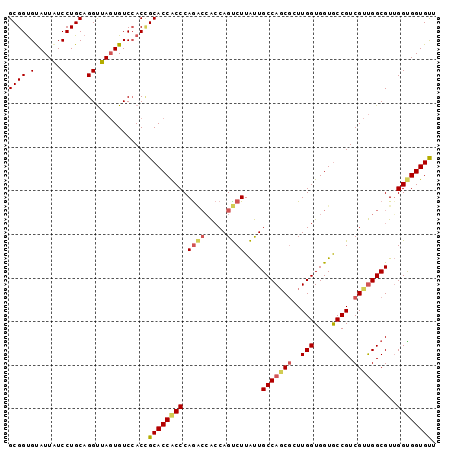
| Location | 7,558,905 – 7,559,003 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -26.96 |
| Energy contribution | -28.63 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7558905 98 + 22224390 GCGGUGUAUUGUCCUGCAGGUUAGUGUCGACGGCACCACCCAGACCACCAGUCUUAUUGCCAGCCGUUGGUGGUGCCGUCGUUGGCGUUGGCGGUGUU ((((.(......)))))..(((((((((((((((((((((.((((.....))))....(....)....)))))))))))))...))))))))...... ( -36.50) >DroEre_CAF1 62891 98 + 1 GCGGCGUAUUAUCCUGCAGGUUAGUGUCCACCGCACCACCCAGGCCACCAGCCUUGUUGCCACCGUUUGGUGGUACCGUCGUUGGCGUGGGUGGUGCU ((((.(......))))).(((........)))((((((((((.((((...((..((.(((((((....))))))).))..)))))).)))))))))). ( -48.80) >DroYak_CAF1 52889 98 + 1 GCGGUGCAUUAUCCUGCAGGCUACUGUCCACUGCACCGCCCAAUGAGGCAGUCUUACUGCCAGCGCUUGGUGGUGCCGUCGCCGGCGUUGGUGGUGUU ((((((((.......((((....))))....))))))))...............(((..((((((((.(((((.....)))))))))))))..))).. ( -43.10) >consensus GCGGUGUAUUAUCCUGCAGGUUAGUGUCCACCGCACCACCCAGACCACCAGUCUUAUUGCCAGCGCUUGGUGGUGCCGUCGUUGGCGUUGGUGGUGUU ((((.(......)))))...............((((((((.((((.....))))....(((((((..(((.....))).)))))))...)))))))). (-26.96 = -28.63 + 1.67)

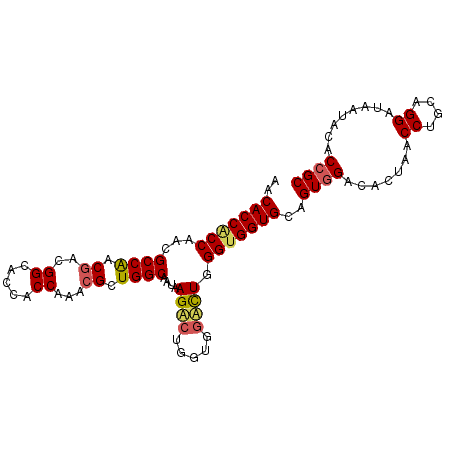
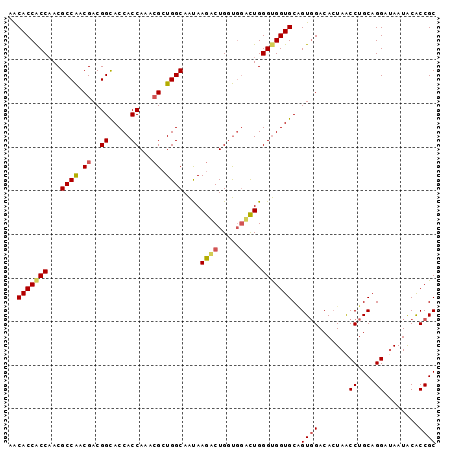
| Location | 7,558,905 – 7,559,003 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -22.20 |
| Energy contribution | -23.09 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7558905 98 - 22224390 AACACCGCCAACGCCAACGACGGCACCACCAACGGCUGGCAAUAAGACUGGUGGUCUGGGUGGUGCCGUCGACACUAACCUGCAGGACAAUACACCGC ....((((....))...(((((((((((((.((.((..(........)..)).))...))))))))))))).............))............ ( -38.00) >DroEre_CAF1 62891 98 - 1 AGCACCACCCACGCCAACGACGGUACCACCAAACGGUGGCAACAAGGCUGGUGGCCUGGGUGGUGCGGUGGACACUAACCUGCAGGAUAAUACGCCGC .((((((((((.((((....((((.(((((....))))).......)))).)))).))))))))))((((.....((.((....)).))...)))).. ( -45.60) >DroYak_CAF1 52889 98 - 1 AACACCACCAACGCCGGCGACGGCACCACCAAGCGCUGGCAGUAAGACUGCCUCAUUGGGCGGUGCAGUGGACAGUAGCCUGCAGGAUAAUGCACCGC ............((((....))))....((((.....(((((.....)))))...))))((((((((.((..(.((.....)).)..)).)))))))) ( -38.10) >consensus AACACCACCAACGCCAACGACGGCACCACCAAACGCUGGCAAUAAGACUGGUGGACUGGGUGGUGCAGUGGACACUAACCUGCAGGAUAAUACACCGC ..(((((((...((((.((..((.....))...)).))))....((((.....)))).)))))))..((((.......((....))........)))) (-22.20 = -23.09 + 0.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:38 2006