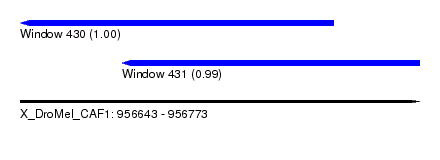
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 956,643 – 956,773 |
| Length | 130 |
| Max. P | 0.996126 |

| Location | 956,643 – 956,745 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -27.95 |
| Energy contribution | -29.40 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 956643 102 - 22224390 --------ACUCGUAACUGGUAACUGGUGGCUGGUAACUGGUAACUGGUAACCGGUAACCGGUAACAGUUAACUGGGUGCAGUCACUGAAUCCGAAAGGGUUGGCUAUGG --------....(((.(..(((((((...((((((.((((((........)))))).))))))..))))).))..).)))(((((....((((....))))))))).... ( -40.60) >DroEre_CAF1 48020 88 - 1 --------ACUCGUAACUGGUGAC--------------UGGUAACUGGCAACUGGUAGCCGGUAACAGUUAACUGGGUGCAGUCACUGAAUCCGCAAGGGUUGGCUAUGG --------...((((.((((((((--------------((...((((((........))))))..(((....)))....)))))))).(((((....))))))).)))). ( -33.20) >DroYak_CAF1 42578 110 - 1 AACUCGCAACUCGUAACUGGUGACUGGUGACUGGUUACUGGUAACUGGUAACUGGUAGUCGGUAACAGUUAACUGGGUGCAGUGACUGAAUCCGAAGGGGUUGGCUAUGG .(((.(((.(((.((((((...(((((..((..(((((..(...)..)))))..))..)))))..))))))...))))))))).....(((((....)))))........ ( -39.90) >consensus ________ACUCGUAACUGGUGACUGGUG_CUGGU_ACUGGUAACUGGUAACUGGUAGCCGGUAACAGUUAACUGGGUGCAGUCACUGAAUCCGAAAGGGUUGGCUAUGG ............((.((..((.((((((((....)))))))).))..)).))(.((((((.....((((..((((....)))).))))(((((....))))))))))).) (-27.95 = -29.40 + 1.45)

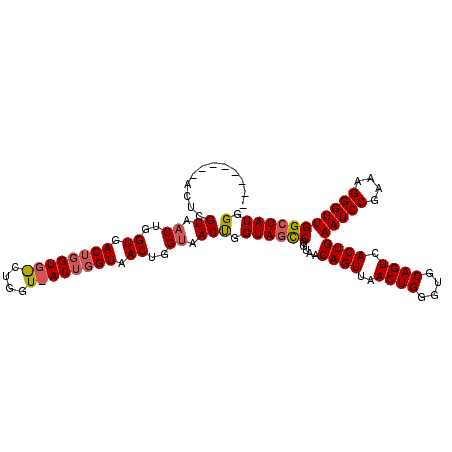
| Location | 956,676 – 956,773 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.59 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -18.32 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 956676 97 - 22224390 UCCCCUCUCUUCUCCCCCGUCAACUGUA--------------ACUCGUAACUGGUAACUGGUGGCUGGUAACUGGUAACUGGUAACCGGUAACCGGUAACAGUUAACUGGG ..............(((.((.((((((.--------------(((.((.((((((.((..((.((..(...)..)).))..)).)))))).)).))).)))))).)).))) ( -35.90) >DroEre_CAF1 48053 82 - 1 UCCCCUGCCUCCCGCCCC-CGAGCUGCA--------------ACUCGUAACUGGUGAC--------------UGGUAACUGGCAACUGGUAGCCGGUAACAGUUAACUGGG ..........(((.....-((((.....--------------.))))((((((...((--------------((((.((((....).))).))))))..))))))...))) ( -26.60) >DroYak_CAF1 42611 111 - 1 UUCCCCACCUCCAGCCCCUUGAGUUGCAACUGGCAACUCGCAACUCGUAACUGGUGACUGGUGACUGGUUACUGGUAACUGGUAACUGGUAGUCGGUAACAGUUAACUGGG ..........((((......(((((((....(....)..))))))).((((((...(((((..((..(((((..(...)..)))))..))..)))))..)))))).)))). ( -42.90) >consensus UCCCCUACCUCCAGCCCC_UGAGCUGCA______________ACUCGUAACUGGUGACUGGUG_CUGGU_ACUGGUAACUGGUAACUGGUAGCCGGUAACAGUUAACUGGG ..............(((....(((((.......................((..((.((((((((....)))))))).))..)).(((((...)))))..)))))....))) (-18.32 = -19.00 + 0.68)

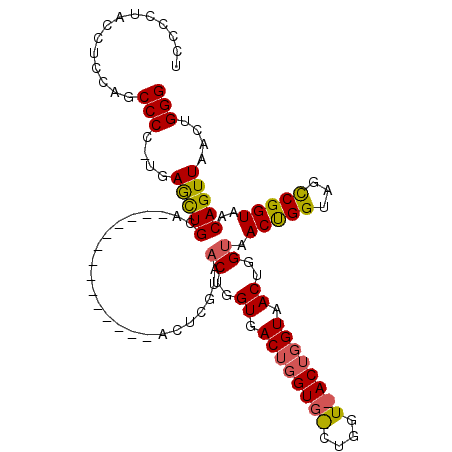
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:47 2006